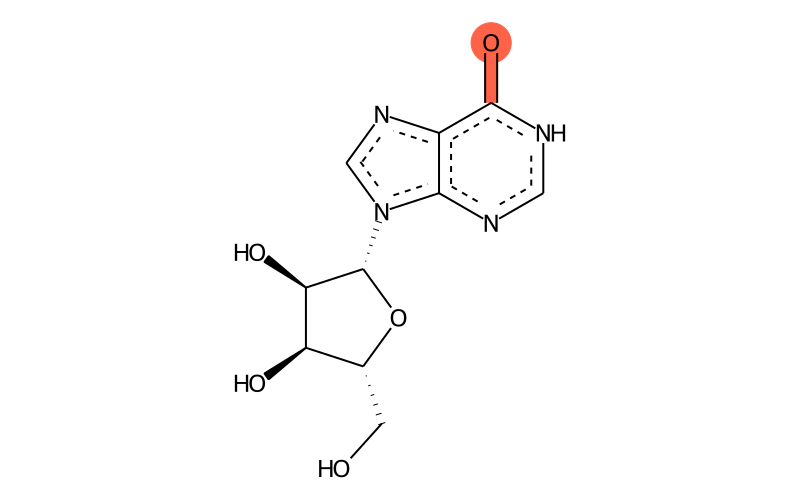
Summary
| Full name | inosine |
| IUPAC name | 9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-1H-purin-6-one |
| Short name | I |
| MODOMICS code new | 2000000009A |
| MODOMICS code | 9A |
| Synonyms |
1,9-dihydro-9-b-D-ribofuranosyl-6H-Purin-6-one
1,9-Dihydro-9-beta-delta-ribofuranosyl-6H-purin-6-one 1,9-Dihydro-9-beta-D-ribofuranosyl-6H-purin-6-one 1a4m 2ada 2fqw (2R,3R,4S,5R)-2-(6-Hydroxy-9H-purin-9-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol (2R,3S,4R,5R)-2-(hydroxymethyl)-5-(6-hydroxy-9H-purin-9-yl)tetrahydrofuran-3,4-diol (2R,3S,4R,5R)-2-(hydroxymethyl)-5-(6-hydroxypurin-9-yl)oxolane-3,4-diol 3h-inosine 58-63-9 5A614L51CT 6H-Purin-6-one, 1,9-dihydro-9-beta-D-ribofuranosyl- 9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-1H-purin-6-one 9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-3H-purin-6-one 9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-6,9-dihydro-1H-purin-6-one 9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-6,9-dihydro-3H-purin-6-one 9-((2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl)-1H-purin-6(9H)-one 9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl]-1H-purin-6-one 9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl]-3H-purin-6-one 9-[(2S,5S)-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl]-1,9-dihydro-6H-purin-6-one 9-(3,4-DIHYDROXY-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-2-YL)-1,9-DIHYDRO-PURIN-6-ONE 9-b-D-ribofuranosyl-Hypoxanthine 9-b-D-Ribofuranosylhypoxanthine 9-beta-delta-ribofuranosyl-Hypoxanthine 9-beta-delta-Ribofuranosylhypoxanthine 9beta-delta-Ribofuranosylhypoxanthine 9-beta-D-Ribofuranosyl-1,9-dihydro-6H-purin-6-one (Inosine) 9-beta-D-ribofuranosyl-9H-purin-6-ol 9-(beta-D-ribofuranosyl)-9H-purin-6-ol 9-beta-D-ribofuranosyl-Hypoxanthine 9-beta-D-Ribofuranosylhypoxanthine 9.beta.-D-Ribofuranosylhypoxanthine 9-.beta.-D-Ribofuranosylhypoxanthine 9beta-D-Ribofuranosylhypoxanthine A-8687 AB0066076 AB1000320 AC1L1LMO AC1Q77T9 AC89F8E4-FD89-45EE-8B41-94DFC07AB42F AI3-52241 AKOS015969695 AKOS015995607 AKOS024462561 AM83935 AMY30713 AN-889 ANW-33000 ARONIS24558 AS-11754 Atorel Atorel; HXR; Hypoxanthine D-riboside B3677 BDBM22104 BDBM50366815 beta-delta-Ribofuranoside hypoxanthine-9 .beta.-D-Ribofuranoside, hypoxanthine-9 beta-D-Ribofuranoside hypoxanthine-9 beta-D-Ribofuranoside, hypoxanthine-9 .beta.-Inosine beta-Inosine bmse000098 bmse000888 bmse000978 BRD-K79612754-001-21-7 C00294 C10-H12-N4-O5 C10H12N4O5 CAS-58-63-9 Catacol CCG-267127 CHEBI:17596 CHEMBL1556 CID 5274258 cid_6021 CID6021 CJ-13587 cMAP_000084 CS-5845 CTK1G9965 D00054 D007288 DB-029916 DB04335 D-Inosine DSSTox_CID_25993 DSSTox_GSID_45993 DSSTox_RID_81278 DTXSID2045993 EINECS 200-390-4 ERK5-6N10 GTPL4554 HMS2235I17 HXR HY-N0092 Hypoxanthine 9-|A-D-ribofuranoside Hypoxanthine 9-beta-delta-ribofuranoside Hypoxanthine 9-beta-D-ribofuranoside Hypoxanthine, 9-beta-D-ribofuranosyl- Hypoxanthine, 9-.beta.-D-ribofuranosyl- Hypoxanthine arabinoside Hypoxanthine D-riboside Hypoxanthine nucleoside Hypoxanthine ribonucleoside Hypoxanthine riboside hypoxanthine-9 beta-delta-Ribofuranoside hypoxanthine-9 beta-D-Ribofuranoside Hypoxanthine-9-beta-delta-ribofuranoside Hypoxanthine-9-beta-D-ribofuranoside Hypoxanthine-9-delta-ribofuranoside Hypoxanthine-9-D-ribofuranoside hypoxanthine-ribose Hypoxanthosine I0037 InChI=1/C10H12N4O5/c15-1-4-6(16)7(17)10(19-4)14-3-13-5-8(14)11-2-12-9(5)18/h2-4,6-7,10,15-17H,1H2,(H,11,12,18)/t4-,6-,7-,10-/m1/s INDOLE-3-CARBOXALDEHYDE INO INO 495 INO-1001 Inosie inosin- Inosin Inosina Inosina [INN-Spanish] inosine (-)-Inosine Inosine- Inosine (8CI,9CI) Inosine, 99% Inosine, 99% - 100G 100g Inosine 99% for biochemistry 5gm Inosine, >=99% (HPLC) Inosine [INN:JAN] Inosine (JAN/INN) Inosine (<lt/> 100 microg/ml) in methanol Inosine,(S) Inosinum Inosinum [INN-Latin] Inotin (TN) iso-prinosine J10406 KBio2_002560 KBio2_005128 KBio2_007696 KBio3_003038 KBioGR_002560 KBioSS_002569 KSC269S6L LS-84000 MCULE-3095132846 MFCD00066770 MLS000028518 NCGC00095787-01 NCGC00095787-05 NCGC00096119-02 NCGC00096119-03 NOS NS00010210 NSC 20262 NSC-20262 NSC20262 Opera_ID_1628 Oxiamin Panholic-L Pantholic-L PubChem14189 Q422564 Ribonosine riboxine RTR-037177 s2442 SCHEMBL15804 Selfer SMP1_000165 SMR000058316 SR-01000721862 SR-01000721862-2 SR-01000721862-3 Tox21_111522 Tox21_111522_1 TR-037177 Trophicardyl UGQMRVRMYYASKQ-KQYNXXCUSA-N UNII-5A614L51CT ZINC8855117 |
| Nature of the modified residue | Natural |
| RNAMods code | I |
| Residue unique ID | 113 |
| Found in RNA | Yes |
| RCSB ligands | |
| Related nucleotides | 217 |
| Enzymes |
ADAR (Homo sapiens) ADARB1 (Homo sapiens) ADAT2 (Homo sapiens) ADAT2 (Trypanosoma cruzi) ADAT2 (Branchiostoma japonicum) ADAT2 (Mus musculus) ADAT3 (Homo sapiens) Tad1 (Saccharomyces cerevisiae) Tad1 (Homo sapiens) Tad2 (Saccharomyces cerevisiae) Tad2 (Fusarium graminearum) Tad3 (Saccharomyces cerevisiae) TadA (Escherichia coli) TadA (Arabidopsis thaliana) |
| Found in phylogeny | Eubacteria, Eukaryota |
| Found naturally in RNA types | tRNA |
Chemical information
* Chemical properties calculated with Open Babel - O'Boyle et al. Open Babel: An open chemical toolbox. J Cheminform 3, 33 (2011) (link)
QM Data:
| Dipole Magnitude [D]: | 9.954064752 |
| Energy [Eh]: | -983.071904535756 |
| HOMO [eV]: | -9.024 |
| LUMO [eV]: | 1.0918 |
| Gap [eV]: | 10.1158 |
Download QM Data:
| Charges | charge.txt |
Download Structures
| 2D | .png .mol .mol2 .sdf .pdb .smi |
| 3D | .mol .mol2 .sdf .pdb |
Tautomers
| Tautomers SMILES |
OCC1C(O)C(O)C(n2c3ncnc(O)c3nc2)O1 tautomer #0
OCC1C(O)C(O)C(n2c3[nH]cnc(=O)c3nc2)O1 tautomer #1 OCC1C(O)C(O)C(n2c3nc[nH]c(=O)c3nc2)O1 tautomer #2 OCC1C(O)C(O)C(n2c3ncnc(O)c3nc2)O1 tautomer #3 OCC1C(O)C(O)C(N2C3=NC=NC(=O)C3N=C2)O1 tautomer #4 |
| Tautomer image | Show Image |
Predicted CYP Metabolic Sites
| CYP3A4 | CYP2D6 | CYP2C9 |
|---|---|---|
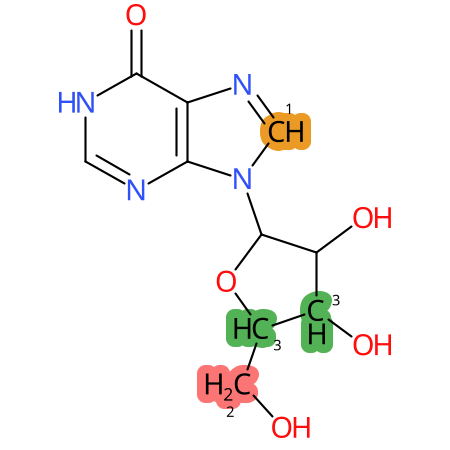
|
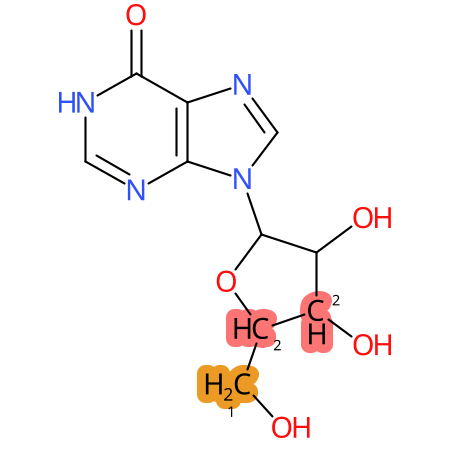
|
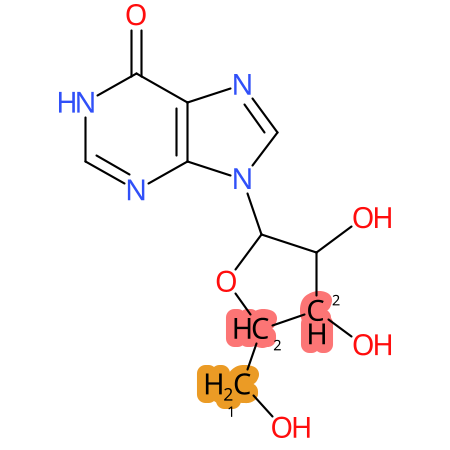
|
* CYP Metabolic sites predicted with SMARTCyp. SMARTCyp is a method for prediction of which sites in a molecule that are most liable to metabolism by Cytochrome P450. It has been shown to be applicable to metabolism by the isoforms 1A2, 2A6, 2B6, 2C8, 2C19, 2E1, and 3A4 (CYP3A4), and specific models for the isoform 2C9 (CYP2C9) and isoform 2D6 (CYP2D6). CYP3A4, CYP2D6, and CYP2C9 are the three of the most important enzymes in drug metabolism since they are involved in the metabolism of more than half of the drugs used today. The three top-ranked atoms are highlighted. See: SmartCYP and SmartCYP - background; Patrik Rydberg, David E. Gloriam, Lars Olsen, The SMARTCyp cytochrome P450 metabolism prediction server, Bioinformatics, Volume 26, Issue 23, 1 December 2010, Pages 2988–2989 (link)
LC-MS Information
| Monoisotopic mass | 268.0808 |
| Average mass | 268.226 |
| [M+H]+ | 269.0886 |
| Product ions | 137 |
| Normalized LC elution time * | 0,95 (Kellner 2014, Kellner 2014) |
| LC elution order/characteristics | between U and G (Kellner 2014, Kellner 2014) |
* normalized to guanosine (G), measured with a RP C-18 column with acetonitrile/ammonium acetate as mobile phase.
LC-MS Publications
| Title | Authors | Journal | Details | ||
|---|---|---|---|---|---|
| Profiling of RNA modifications by multiplexed stable isotope labelling. | Kellner S, Neumann J, Rosenkranz D, Lebedeva S, Ketting RF, Zischler H, Schneider D, Helm M. | Chem Commun (Camb). | [details] | 24567952 | - |
| Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. | Su D, Chan CT, Gu C, Lim KS, Chionh YH, McBee ME, Russell BS, Babu IR, Begley TJ, Dedon PC... | Nat Protoc | [details] | 24625781 | - |
| Absolute and relative quantification of RNA modifications via biosynthetic isotopomers. | Kellner S, Ochel A, Thuring K, Spenkuch F, Neumann J, Sharma S, Entian KD, Schneider D, Helm M... | Nucleic Acids Res | [details] | 25129236 | - |
Chemical groups contained
| Type | Subtype |
|---|---|
| other | keto |
Reactions producing inosine
| Name |
|---|
| A:I |
Reactions starting from inosine
| Name |
|---|
| I:m1I |
| I:Im |
Last modification of this entry: Sept. 15, 2025