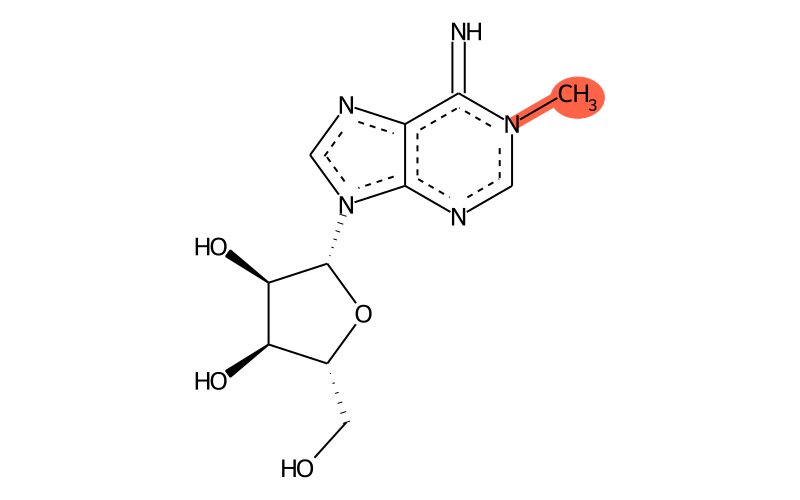
Summary
| Full name | 1-methyladenosine |
| IUPAC name | (2R,3S,4R,5R)-2-(hydroxymethyl)-5-(6-imino-1-methylpurin-9-yl)oxolane-3,4-diol |
| Short name | m1A |
| MODOMICS code new | 2000000001A |
| MODOMICS code | 1A |
| Synonyms |
15763-06-1
1-Methyladenosine 1-Methyladenosine, 98% - 250MG 250mg (2R,3R,4S,5R)-2-(6-Amino-1-methyl-1H-purin-9(2H)-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol (2R,3S,4R,5R)-2-(hydroxymethyl)-5-(6-imino-1-methyl-1H-purin-9(6H)-yl)tetrahydrofuran-3,4-diol (2R,3S,4R,5R)-2-(hydroxymethyl)-5-(6-imino-1-methyl-6,9-dihydro-1H-purin-9-yl)oxolane-3,4-diol (2R,3S,4R,5R)-2-(hydroxymethyl)-5-(6-imino-1-methylpurin-9-yl)oxolane-3,4-diol (2R,3S,4R,5R)-2-(hydroxymethyl)-5-(6-imino-1-methyl-purin-9-yl)tetrahydrofuran-3,4-diol AC1L1DAV AC-32347 Adenosine, 1-methyl- Adenosine, 1-methyl- (VAN) (8CI) Adenosine, N,6-didehydro-1,6-dihydro-1-methyl- Adenosine, N,6-didehydro-1,6-dihydro-1-methyl- (9CI) Adenosine, N,6-didehydro-1,9-dihydro-1-methyl- AKOS030241121 alpha-Vetivone C02494 C11-H15-N5-O4 CHEBI:16020 CHEMBL1866485 CID27476 CS-0059513 CTK8F3395 DTXSID30864632 GFYLSDSUCHVORB-IOSLPCCCSA-N HY-113081 K-4948 M1A MFCD00038421 N1-Methyladenosine N(1)-methyladenosine N1-Methyl-D-adenosine NCGC00163305-01 NS00099498 NSC 92165 Q161643 SCHEMBL19553541 ZINC100053615 |
| Nature of the modified residue | Natural |
| RNAMods code | Ѣ |
| Residue unique ID | 83 |
| Found in RNA | Yes |
| Related nucleotides | 204 |
| Enzymes |
Bmt2 (Saccharomyces cerevisiae) KamB (Streptomyces sp. DSM 40477) NpmA (Escherichia coli) NpmC (Clostridia) Rrp8 (Saccharomyces cerevisiae) TRMT10C (Homo sapiens) Trm10p (Thermococcus kodakaraensis) Trm61 (Saccharomyces cerevisiae) TrmI (Pyrococcus abyssi) TrmI (Thermus thermophilus) TrmI (Mycobacterium tuberculosis) TrmI (Haloferax volcanii) TrmK (Bacillus subtilis) Trmt61A (Homo sapiens) Trmt61B (Homo sapiens) |
| Found in phylogeny | Archaea, Eubacteria, Eukaryota |
| Found naturally in RNA types | rRNA, tRNA |
Chemical information
| Sum formula | C11H15N5O4 |
| Type of moiety | nucleoside |
| Degeneracy | not applicable |
| PubChem ID | 27476 |
| ChEBI ID | 16020 |
| CAS Registry Number | 15763-06-1 |
| Reaxys Registry Number | 42832 |
| SMILES | C[n]1c(=N)c2c([n]([C@H]3[C@H](O)[C@H](O)[C@@H](CO)O3)cn2)nc1 |
| logP | -2.0395 |
| TPSA | 129.41 |
| Number of atoms | 20 |
| Number of Hydrogen Bond Acceptors 1 (HBA1) | 7 |
| Number of Hydrogen Bond Acceptors 2 (HBA2) | 9 |
| Number of Hydrogen Bond Donors (HBD) | 4 |
| InChI | InChI=1S/C11H15N5O4/c1-15-3-14-10-6(9(15)12)13-4-16(10)11-8(19)7(18)5(2-17)20-11/h3-5,7-8,11-12,17-19H,2H2,1H3/t5-,7-,8-,11-/m1/s1 |
| InChIKey | GFYLSDSUCHVORB-IOSLPCCCSA-N |
| Search the molecule in external databases | ChEMBL PubChem Compound Database Ligand Expo WIPO |
| PubChem CID | |
| PubChem SIDs |
5506
610755 8145039 8169807 17171932 26756660 34669998 40649335 57310453 57391286 76691120 104300100 134992420 163128306 185986534 189415457 242462149 250031422 254789874 268755350 273528948 304755946 310278077 312226601 318050922 319081829 319441218 337257024 341241399 347244718 348276024 348804347 354067038 363603225 374040679 375079201 375097323 375987055 375987306 378034043 381012308 381930085 384441081 385644741 386481073 403479323 404622946 404757870 406572785 406804801 419589687 433774864 433775410 433777103 433777104 433777105 433777703 434413348 439458983 440703496 |
* Chemical properties calculated with Open Babel - O'Boyle et al. Open Babel: An open chemical toolbox. J Cheminform 3, 33 (2011) (link)
QM Data:
| Dipole Magnitude [D]: | 8.174864284 |
| Energy [Eh]: | -1002.49687613282 |
| HOMO [eV]: | -8.4504 |
| LUMO [eV]: | 1.1952 |
| Gap [eV]: | 9.6456 |
Download QM Data:
| Charges | charge.txt |
Download Structures
| 2D | .png .mol .mol2 .sdf .pdb .smi |
| 3D | .mol .mol2 .sdf .pdb |
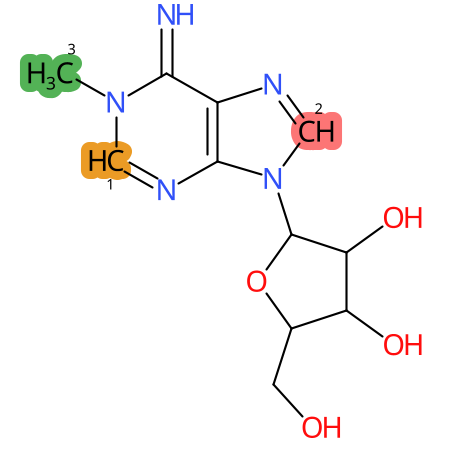
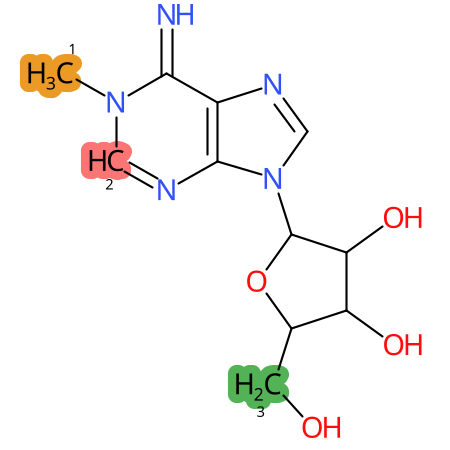
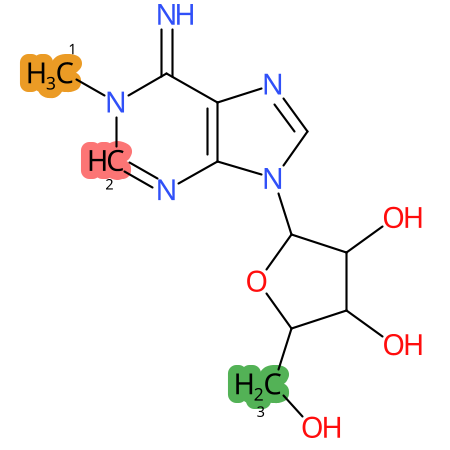
Predicted CYP Metabolic Sites
| CYP3A4 | CYP2D6 | CYP2C9 |
|---|---|---|
|
|
|
* CYP Metabolic sites predicted with SMARTCyp. SMARTCyp is a method for prediction of which sites in a molecule that are most liable to metabolism by Cytochrome P450. It has been shown to be applicable to metabolism by the isoforms 1A2, 2A6, 2B6, 2C8, 2C19, 2E1, and 3A4 (CYP3A4), and specific models for the isoform 2C9 (CYP2C9) and isoform 2D6 (CYP2D6). CYP3A4, CYP2D6, and CYP2C9 are the three of the most important enzymes in drug metabolism since they are involved in the metabolism of more than half of the drugs used today. The three top-ranked atoms are highlighted. See: SmartCYP and SmartCYP - background; Patrik Rydberg, David E. Gloriam, Lars Olsen, The SMARTCyp cytochrome P450 metabolism prediction server, Bioinformatics, Volume 26, Issue 23, 1 December 2010, Pages 2988–2989 (link)
LC-MS Information
| Monoisotopic mass | 281.1124 |
| Average mass | 281.268 |
| [M+H]+ | 282.1202 |
| Product ions | 150 |
| Normalized LC elution time * | 0,64 (Kellner 2014); 0,72 (Kellner 2014) |
| LC elution order/characteristics | between U and G (Kellner 2014, Kellner 2014) |
* normalized to guanosine (G), measured with a RP C-18 column with acetonitrile/ammonium acetate as mobile phase.
LC-MS Publications
| Title | Authors | Journal | Details | ||
|---|---|---|---|---|---|
| Profiling of RNA modifications by multiplexed stable isotope labelling. | Kellner S, Neumann J, Rosenkranz D, Lebedeva S, Ketting RF, Zischler H, Schneider D, Helm M. | Chem Commun (Camb). | [details] | 24567952 | - |
| Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. | Su D, Chan CT, Gu C, Lim KS, Chionh YH, McBee ME, Russell BS, Babu IR, Begley TJ, Dedon PC... | Nat Protoc | [details] | 24625781 | - |
| Absolute and relative quantification of RNA modifications via biosynthetic isotopomers. | Kellner S, Ochel A, Thuring K, Spenkuch F, Neumann J, Sharma S, Entian KD, Schneider D, Helm M... | Nucleic Acids Res | [details] | 25129236 | - |
Comments
m1A has a positive charge at neutral pH.
Chemical groups contained
| Type | Subtype |
|---|---|
| methyl group | methyl at aromatic N |
Reactions producing 1-methyladenosine
| Name |
|---|
| A:m1A |
Reactions starting from 1-methyladenosine
| Name |
|---|
| m1A:m1I |
| m1A:m1Am |
| m1A:A |
Last modification of this entry: Sept. 15, 2025