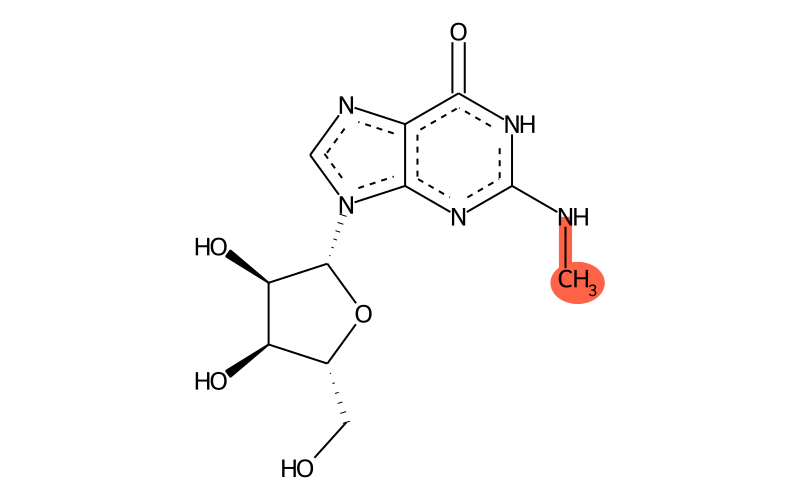
Summary
| Full name | N2-methylguanosine |
| IUPAC name | 9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-2-(methylamino)-1H-purin-6-one |
| Short name | m2G |
| MODOMICS code new | 2000000002G |
| MODOMICS code | 2G |
| Synonyms |
2140-77-4
2-(methylamino)-9-(beta-D-ribofuranosyl)-1,9-dihydro-6H-purin-6-one 2-Methylguanosine 2-(Methylimino)-9-pentofuranosyl-3,9-dihydro-2H-purin-6-ol 45T7B5IAN4 7-methylguanosine 9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-2-(methylamino)-1H-purin-6-one 9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-2-(methylamino)-3H-purin-6-one 9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-2-(methylamino)-6,9-dihydro-3H-purin-6-one 9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl]-2-(methylamino)-1,9-dihydro-6H-purin-6-one 9-((2R,3R,4S,5R)-3,4-Dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl)-2-(methylamino)-1H-purin-6(9H)-one AC1MHZC4 CHEBI:19702 CHEMBL3351027 CID3035422 CS-0089200 CTK1A0934 DTXSID20943960 Guanosine, N-methyl- HY-111647 J-014035 m2g MFCD00057039 N2-Methylguanine riboside N2-Methylguanosine N(2)-Methylguanosine N/A N-Methyl guanosine n-methyl-Guanosine N-methylguanosine NS00014684 Q1419713 SCHEMBL41509 SLEHROROQDYRAW-KQYNXXCUSA-N UNII-45T7B5IAN4 ZINC12496640 |
| Nature of the modified residue | Natural |
| RNAMods code | L |
| Residue unique ID | 36 |
| Found in RNA | Yes |
| Related nucleotides | 191 |
| Enzymes |
RlmG (Escherichia coli) RlmKL (Escherichia coli) RsmC (Escherichia coli) RsmD (Escherichia coli) RsmD (Arabidopsis thaliana) RsmJ (Escherichia coli) THUMPD2 (Homo sapiens) THUMPD3 (Homo sapiens) TRM11 (Homo sapiens) TRM112 (Homo sapiens) Trm-G10 (Pyrococcus abyssi) Trm1 (Saccharomyces cerevisiae) Trm1 (Aquifex aeolicus) Trm1 (Pyrococcus horikoshii) Trm1 (Homo sapiens) Trm11 (Saccharomyces cerevisiae) TrmN (Methanocaldococcus jannaschii) TrmN (Thermus thermophilus) trm1 (Haloferax volcanii) trmG10 (Haloferax volcanii) |
| Found in phylogeny | Archaea, Eubacteria, Eukaryota |
| Found naturally in RNA types | rRNA, snRNA, tRNA |
Chemical information
| Sum formula | C11H15N5O5 |
| Type of moiety | nucleoside |
| Degeneracy | not applicable |
| PubChem ID | 3035422 |
| ChEBI ID | 19702 |
| CAS Registry Number | 2140-77-4 |
| Reaxys Registry Number | 46491 1031383 |
| SMILES | CNc1[nH]c(=O)c2nc[n]([C@@H]3O[C@H](CO)[C@@H](O)[C@H]3O)c2n1 |
| logP | -2.1542 |
| TPSA | 145.52 |
| Number of atoms | 21 |
| Number of Hydrogen Bond Acceptors 1 (HBA1) | 8 |
| Number of Hydrogen Bond Acceptors 2 (HBA2) | 9 |
| Number of Hydrogen Bond Donors (HBD) | 5 |
| InChI | InChI=1S/C11H15N5O5/c1-12-11-14-8-5(9(20)15-11)13-3-16(8)10-7(19)6(18)4(2-17)21-10/h3-4,6-7,10,17-19H,2H2,1H3,(H2,12,14,15,20)/t4-,6-,7-,10-/m1/s1 |
| InChIKey | SLEHROROQDYRAW-KQYNXXCUSA-N |
| Search the molecule in external databases | ChEMBL PubChem Compound Database Ligand Expo WIPO |
| PubChem CID | |
| PubChem SIDs |
16951281
36078900 53801189 57353088 75014986 111638931 129252284 135078236 162495704 163536797 226426524 241043531 249896385 252354222 252400676 254770362 259291005 273528944 310278081 312226901 312366515 332868903 341232526 348276777 349717712 363671325 375091219 375300378 377991387 381833205 381833428 384501010 385020711 385532187 385641751 386478018 387154019 389159813 404769320 419580121 433775053 433775452 434369797 434407454 441514789 442031544 443531232 |
* Chemical properties calculated with Open Babel - O'Boyle et al. Open Babel: An open chemical toolbox. J Cheminform 3, 33 (2011) (link)
QM Data:
| Dipole Magnitude [D]: | 10.397238988 |
| Energy [Eh]: | -1077.72840328574 |
| HOMO [eV]: | -8.3381 |
| LUMO [eV]: | 1.3896 |
| Gap [eV]: | 9.7277 |
Download QM Data:
| Charges | charge.txt |
Download Structures
| 2D | .png .mol .mol2 .sdf .pdb .smi |
| 3D | .mol .mol2 .sdf .pdb |
Tautomers
| Tautomers SMILES |
CN=c1[nH]c(=O)c2ncn(C3OC(CO)C(O)C3O)c2[nH]1 tautomer #0
CNc1nc(O)c2ncn(C3OC(CO)C(O)C3O)c2n1 tautomer #1 CNc1nc(=O)c2ncn(C3OC(CO)C(O)C3O)c2[nH]1 tautomer #2 CNc1[nH]c(=O)c2ncn(C3OC(CO)C(O)C3O)c2n1 tautomer #3 CN=c1[nH]c(O)c2ncn(C3OC(CO)C(O)C3O)c2n1 tautomer #4 CN=C1NC(=O)C2N=CN(C3OC(CO)C(O)C3O)C2=N1 tautomer #5 CNc1nc(O)c2ncn(C3OC(CO)C(O)C3O)c2n1 tautomer #6 CNC1=NC(=O)C2N=CN(C3OC(CO)C(O)C3O)C2=N1 tautomer #7 CN=c1nc(O)c2ncn(C3OC(CO)C(O)C3O)c2[nH]1 tautomer #8 CN=C1N=C(O)C2N=CN(C3OC(CO)C(O)C3O)C2=N1 tautomer #9 |
| Tautomer image | Show Image |
Predicted CYP Metabolic Sites
| CYP3A4 | CYP2D6 | CYP2C9 |
|---|---|---|
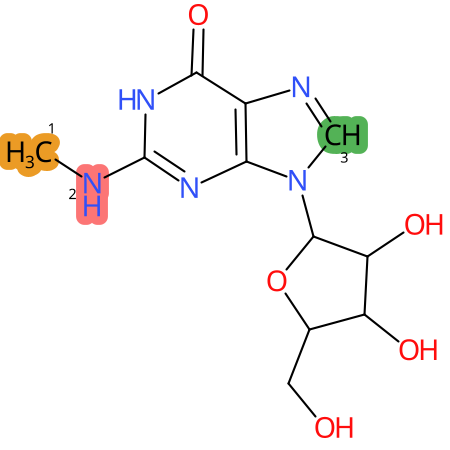
|

|
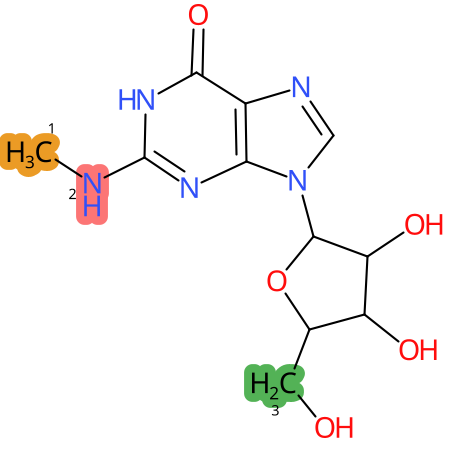
|
* CYP Metabolic sites predicted with SMARTCyp. SMARTCyp is a method for prediction of which sites in a molecule that are most liable to metabolism by Cytochrome P450. It has been shown to be applicable to metabolism by the isoforms 1A2, 2A6, 2B6, 2C8, 2C19, 2E1, and 3A4 (CYP3A4), and specific models for the isoform 2C9 (CYP2C9) and isoform 2D6 (CYP2D6). CYP3A4, CYP2D6, and CYP2C9 are the three of the most important enzymes in drug metabolism since they are involved in the metabolism of more than half of the drugs used today. The three top-ranked atoms are highlighted. See: SmartCYP and SmartCYP - background; Patrik Rydberg, David E. Gloriam, Lars Olsen, The SMARTCyp cytochrome P450 metabolism prediction server, Bioinformatics, Volume 26, Issue 23, 1 December 2010, Pages 2988–2989 (link)
LC-MS Information
| Monoisotopic mass | 297.1073 |
| Average mass | 297.267 |
| [M+H]+ | 298.1151 |
| Product ions | 166 |
| Normalized LC elution time * | not available |
| LC elution order/characteristics | not available |
* normalized to guanosine (G), measured with a RP C-18 column with acetonitrile/ammonium acetate as mobile phase.
Chemical groups contained
| Type | Subtype |
|---|---|
| methyl group | methyl at other N |
Reactions producing N2-methylguanosine
| Name |
|---|
| G:m2G |
Reactions starting from N2-methylguanosine
| Name |
|---|
| m2G:m2,2G |
| m2G:m2Gm |
Last modification of this entry: Sept. 15, 2025