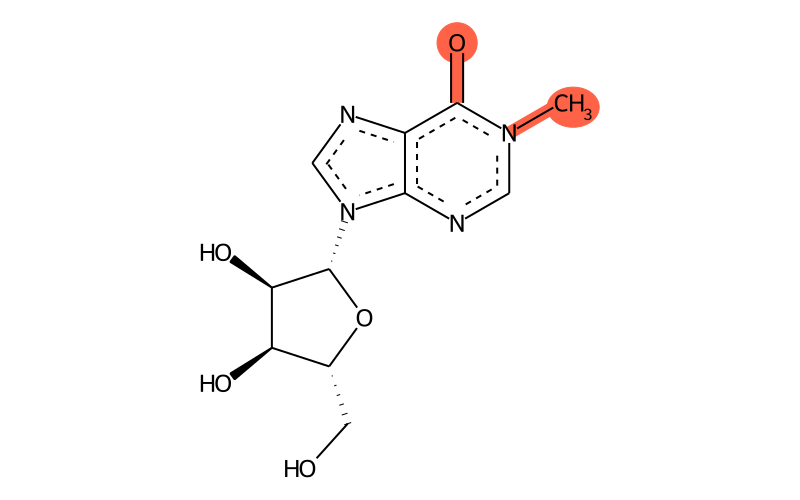
Summary
| Full name | 1-methylinosine |
| IUPAC name | 9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-1-methylpurin-6-one |
| Short name | m1I |
| MODOMICS code new | 2000000019A |
| MODOMICS code | 19A |
| Synonyms |
1-methyl-inosine
1-Methylinosine 2140-73-0 9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-1-methyl-6,9-dihydro-1H-purin-6-one 9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-1-methylpurin-6-one 9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl]-1-methyl-1,9-dihydro-6H-purin-6-one AC1L22KY C6JR3HAB96 CHEBI:19065 CID65095 CS-0061842 DTXSID90175670 HY-113139 inosine, 1-methyl Inosine, 1-methyl- Inosine,1-methyl- m(1)i N1-Methylinosine Q27109107 SCHEMBL19359092 UNII-C6JR3HAB96 WJNGQIYEQLPJMN-IOSLPCCCSA-N |
| Nature of the modified residue | Natural |
| RNAMods code | O |
| Residue unique ID | 26 |
| Found in RNA | Yes |
| Related nucleotides | 344 |
| Enzymes |
Trm5 (Saccharomyces cerevisiae) HVO_2747 (Haloferax volcanii) |
| Found in phylogeny | Eukaryota |
| Found naturally in RNA types | tRNA |
Chemical information
| Sum formula | C11H14N4O5 |
| Type of moiety | nucleoside |
| Degeneracy | not applicable |
| PubChem ID | 65095 |
| ChEBI ID | 19065 |
| CAS Registry Number | 2140-73-0 |
| Reaxys Registry Number | 42831 |
| SMILES | C[n]1c(=O)c2c([n]([C@H]3[C@H](O)[C@H](O)[C@@H](CO)O3)cn2)nc1 |
| logP | -2.2585 |
| TPSA | 122.63 |
| Number of atoms | 20 |
| Number of Hydrogen Bond Acceptors 1 (HBA1) | 7 |
| Number of Hydrogen Bond Acceptors 2 (HBA2) | 9 |
| Number of Hydrogen Bond Donors (HBD) | 3 |
| InChI | InChI=1S/C11H14N4O5/c1-14-3-13-9-6(10(14)19)12-4-15(9)11-8(18)7(17)5(2-16)20-11/h3-5,7-8,11,16-18H,2H2,1H3/t5-,7-,8-,11-/m1/s1 |
| InChIKey | WJNGQIYEQLPJMN-IOSLPCCCSA-N |
| Search the molecule in external databases | ChEMBL PubChem Compound Database Ligand Expo WIPO |
| PubChem CID | |
| PubChem SIDs |
8189507
14824513 43121935 57315309 57392306 76969288 104332863 135022177 135610188 250032172 273525258 312226528 315736069 341161597 342526846 344390272 347744791 348275874 349717705 355073652 381833233 381833456 385527342 385663804 386500320 388608285 404623001 404825304 419580331 434370157 434737980 439432544 442069981 442910328 442932206 |
* Chemical properties calculated with Open Babel - O'Boyle et al. Open Babel: An open chemical toolbox. J Cheminform 3, 33 (2011) (link)
QM Data:
| Dipole Magnitude [D]: | 8.654338289 |
| Energy [Eh]: | -1022.37079326161 |
| HOMO [eV]: | -9.0062 |
| LUMO [eV]: | 1.0625 |
| Gap [eV]: | 10.0687 |
Download QM Data:
| Charges | charge.txt |
Download Structures
| 2D | .png .mol .mol2 .sdf .pdb .smi |
| 3D | .mol .mol2 .sdf .pdb |
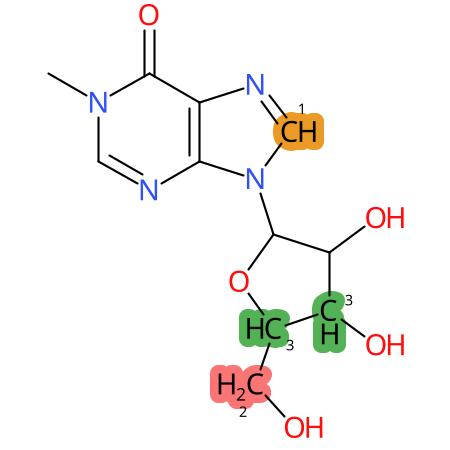
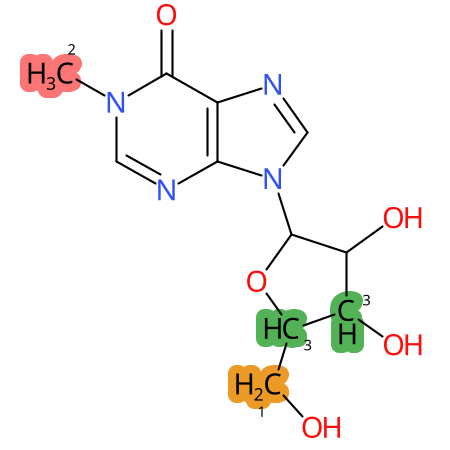
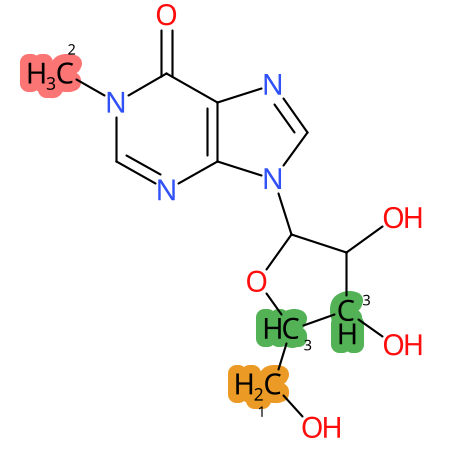
Predicted CYP Metabolic Sites
| CYP3A4 | CYP2D6 | CYP2C9 |
|---|---|---|
|
|
|
* CYP Metabolic sites predicted with SMARTCyp. SMARTCyp is a method for prediction of which sites in a molecule that are most liable to metabolism by Cytochrome P450. It has been shown to be applicable to metabolism by the isoforms 1A2, 2A6, 2B6, 2C8, 2C19, 2E1, and 3A4 (CYP3A4), and specific models for the isoform 2C9 (CYP2C9) and isoform 2D6 (CYP2D6). CYP3A4, CYP2D6, and CYP2C9 are the three of the most important enzymes in drug metabolism since they are involved in the metabolism of more than half of the drugs used today. The three top-ranked atoms are highlighted. See: SmartCYP and SmartCYP - background; Patrik Rydberg, David E. Gloriam, Lars Olsen, The SMARTCyp cytochrome P450 metabolism prediction server, Bioinformatics, Volume 26, Issue 23, 1 December 2010, Pages 2988–2989 (link)
LC-MS Information
| Monoisotopic mass | 282.0964 |
| Average mass | 282.253 |
| [M+H]+ | 283.1042 |
| Product ions | 151 |
| Normalized LC elution time * | 1,24 (Kellner 2014) |
| LC elution order/characteristics | between G and A (Kellner 2014) |
* normalized to guanosine (G), measured with a RP C-18 column with acetonitrile/ammonium acetate as mobile phase.
LC-MS Publications
| Title | Authors | Journal | Details | ||
|---|---|---|---|---|---|
| Profiling of RNA modifications by multiplexed stable isotope labelling. | Kellner S, Neumann J, Rosenkranz D, Lebedeva S, Ketting RF, Zischler H, Schneider D, Helm M. | Chem Commun (Camb). | [details] | 24567952 | - |
| Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. | Su D, Chan CT, Gu C, Lim KS, Chionh YH, McBee ME, Russell BS, Babu IR, Begley TJ, Dedon PC... | Nat Protoc | [details] | 24625781 | - |
Chemical groups contained
| Type | Subtype |
|---|---|
| methyl group | methyl at aromatic N |
| other | keto |
Reactions producing 1-methylinosine
| Name |
|---|
| I:m1I |
| m1A:m1I |
Reactions starting from 1-methylinosine
| Name |
|---|
| m1I:m1Im |
Last modification of this entry: Sept. 15, 2025