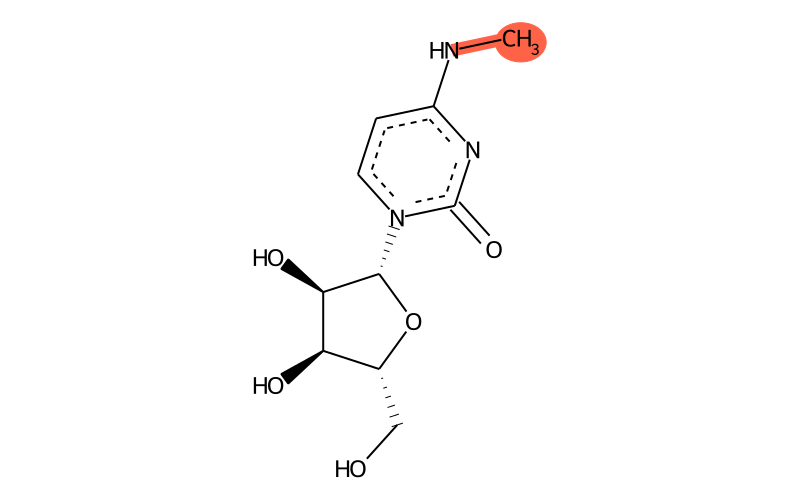
Summary
| Full name | N4-methylcytidine |
| IUPAC name | 1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-4-(methylamino)pyrimidin-2-one |
| Short name | m4C |
| MODOMICS code new | 2000000004C |
| MODOMICS code | 4C |
| Synonyms |
10578-79-7
106542-80-7 1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-4-(methylamino)pyrimidin-2-one 1-((2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl)-4-(methylamino)pyrimidin-2(1H)-one 1-((2R,3S,4R,5S)-3,4-dihydroxy-5-(hydroxymethyl)-tetrahydrofuran-2-yl)-4-(methylamino)pyrimidin-2(1H)-one 1303AA 1-[3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-4-(methylamino)pyrimidin-2-one 4-Methylcytidine AC1L2QU4 AC1Q6C4G AC-32336 AJ-55410 AKOS016011534 AR-1I3429 C10-H15-N3-O5 C10H15N3O5 CID101436 CTK0H8883 Cytidine, N-methyl- DS-6291 DTXSID20147340 MFCD23701766 N-4-Methylcytidine N4-Methylcytidine N(4)-Methylcytidine n-methylcytidine NSC 518744 NSC518744 SCHEMBL157772 ST24048770 ZINC5996572 |
| Nature of the modified residue | Natural |
| RNAMods code | ν |
| Residue unique ID | 9 |
| Found in RNA | Yes |
| Related nucleotides | 281 |
| Enzymes |
CMAL (Arabidopsis thaliana) RsmH (Escherichia coli) |
| Found in phylogeny | Eubacteria |
| Found naturally in RNA types | rRNA |
Chemical information
| Sum formula | C10H15N3O5 |
| Type of moiety | nucleoside |
| Degeneracy | not applicable |
| SMILES | CNc1nc(=O)[n]([C@H]2[C@H](O)[C@H](O)[C@@H](CO)O2)cc1 |
| logP | -2.0305 |
| TPSA | 116.84 |
| Number of atoms | 18 |
| Number of Hydrogen Bond Acceptors 1 (HBA1) | 7 |
| Number of Hydrogen Bond Acceptors 2 (HBA2) | 8 |
| Number of Hydrogen Bond Donors (HBD) | 4 |
| PDB no exac match , link to the most similar ligand | AR3 |
| HMDB (Human Metabolome Database) no exact match, link to the most similar ligand | None |
| InChI | InChI=1S/C10H15N3O5/c1-11-6-2-3-13(10(17)12-6)9-8(16)7(15)5(4-14)18-9/h2-3,5,7-9,14-16H,4H2,1H3,(H,11,12,17)/t5-,7-,8-,9-/m1/s1 |
| InChIKey | LZCNWAXLJWBRJE-ZOQUXTDFSA-N |
| Search the molecule in external databases | ChEMBL ChemAgora ChEBI PubChem Compound Database Ligand Expo ChemSpider WIPO |
| PubChem CID | |
| PubChem SIDs |
10232043
15070134 44431467 57337379 74642989 103816393 104364297 117588129 128562131 135059603 140033892 143178791 162474997 163850446 171579027 189053749 223564581 226521601 249867859 250217078 252228354 252414863 252464702 255444791 257894762 274046820 275926161 297453032 307081829 312605608 315670145 315707950 319134781 319460292 321932182 340570025 341274099 342526824 342526825 346698310 348530313 354314538 355182536 355207376 364121860 374092619 374370630 375360528 376040223 377419311 381990130 383452050 388968399 404823486 406677383 424592044 434696420 436395698 439450949 440684228 440703507 441236939 443498304 |
* Chemical properties calculated with Open Babel - O'Boyle et al. Open Babel: An open chemical toolbox. J Cheminform 3, 33 (2011) (link)
Download Structures
| 2D | .png .mol .mol2 .sdf .pdb .smi |
| 3D | .mol .mol2 .sdf .pdb |
Tautomers
| Tautomers SMILES |
CN=c1[nH]c(=O)n(C2C(O)C(O)C(CO)O2)cc1 tautomer #0
CNc1nc(=O)n(C2C(O)C(O)C(CO)O2)cc1 tautomer #1 CN=c1nc(O)n(C2C(O)C(O)C(CO)O2)cc1 tautomer #2 |
| Tautomer image | Show Image |
Predicted CYP Metabolic Sites
| CYP3A4 | CYP2D6 | CYP2C9 |
|---|---|---|
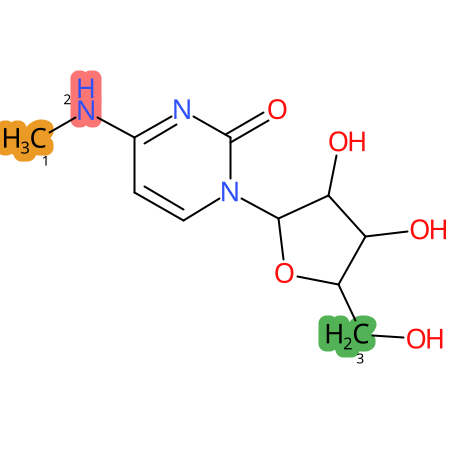
|
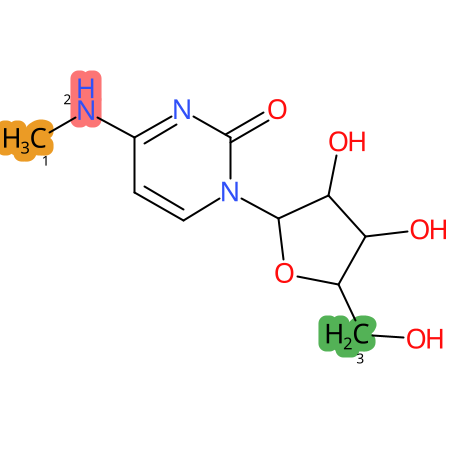
|
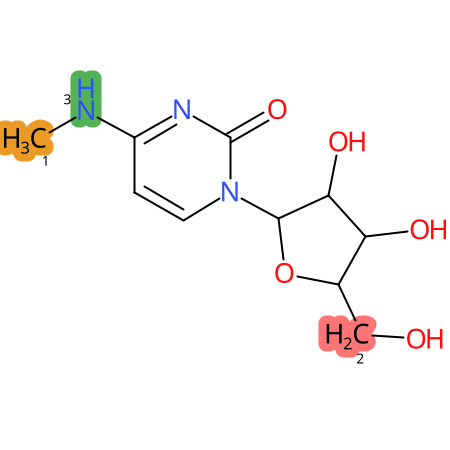
|
* CYP Metabolic sites predicted with SMARTCyp. SMARTCyp is a method for prediction of which sites in a molecule that are most liable to metabolism by Cytochrome P450. It has been shown to be applicable to metabolism by the isoforms 1A2, 2A6, 2B6, 2C8, 2C19, 2E1, and 3A4 (CYP3A4), and specific models for the isoform 2C9 (CYP2C9) and isoform 2D6 (CYP2D6). CYP3A4, CYP2D6, and CYP2C9 are the three of the most important enzymes in drug metabolism since they are involved in the metabolism of more than half of the drugs used today. The three top-ranked atoms are highlighted. See: SmartCYP and SmartCYP - background; Patrik Rydberg, David E. Gloriam, Lars Olsen, The SMARTCyp cytochrome P450 metabolism prediction server, Bioinformatics, Volume 26, Issue 23, 1 December 2010, Pages 2988–2989 (link)
LC-MS Information
| Monoisotopic mass | 257.1012 |
| Average mass | 257.243 |
| [M+H]+ | 258.109 |
| Product ions | 126 |
| Normalized LC elution time * | not available |
| LC elution order/characteristics | not available |
* normalized to guanosine (G), measured with a RP C-18 column with acetonitrile/ammonium acetate as mobile phase.
Chemical groups contained
| Type | Subtype |
|---|---|
| methyl group | methyl at other N |
Reactions producing N4-methylcytidine
| Name |
|---|
| C:m4C |
Reactions starting from N4-methylcytidine
| Name |
|---|
| m4C:m4Cm |
| m4C:m4,4C |
Last modification of this entry: Sept. 22, 2023