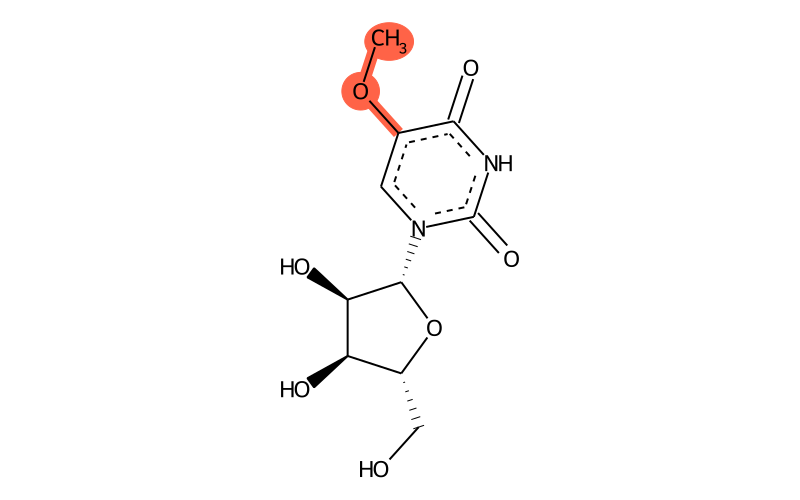
Summary
| Full name | 5-methoxyuridine |
| IUPAC name | 1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-5-methoxypyrimidine-2,4-dione |
| Short name | mo5U |
| MODOMICS code new | 2000000501U |
| MODOMICS code | 501U |
| Synonyms |
1-[(2R,3R,4R,5R)-3,4-Dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-5-methoxypyrimidine-2,4-dione
1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-5-methoxypyrimidine-2,4-dione 1-((2R,3R,4S,5R)-3,4-Dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl)-5-methoxypyrimidine-2,4(1H,3H)-dione 35542-01-9 5-Methoxyuridine AC1LOMX5 AC-32337 AKOS025146635 C10-H14-N2-O7 C10H14N2O7 CHEBI:20601 CID1265899 CTK1C5565 EINECS 252-609-8 MFCD00006533 mo(5)u NS00059240 SCHEMBL41141 Uridine, 5-methoxy- Uridine,5-methoxy- ZB015843 ZINC1078619 |
| Nature of the modified residue | Natural |
| RNAMods code | 5 |
| Residue unique ID | 74 |
| Found in RNA | Yes |
| Related nucleotides | 363 |
| Enzymes |
CmoB (Escherichia coli) TrmR (Bacillus subtilis) |
| Found in phylogeny | Eubacteria |
| Found naturally in RNA types | tRNA |
Chemical information
| Sum formula | C10H14N2O7 |
| Type of moiety | nucleoside |
| Degeneracy | not applicable |
| PubChem ID | 169704 |
| ChEBI ID | 20601 |
| CAS Registry Number | 35542-01-9 |
| Reaxys Registry Number | 894316 |
| SMILES | COc1c(=O)[nH]c(=O)[n]([C@H]2[C@H](O)[C@H](O)[C@@H](CO)O2)c1 |
| logP | -2.8433 |
| TPSA | 134.01 |
| Number of atoms | 19 |
| Number of Hydrogen Bond Acceptors 1 (HBA1) | 7 |
| Number of Hydrogen Bond Acceptors 2 (HBA2) | 8 |
| Number of Hydrogen Bond Donors (HBD) | 4 |
| InChI | InChI=1S/C10H14N2O7/c1-18-4-2-12(10(17)11-8(4)16)9-7(15)6(14)5(3-13)19-9/h2,5-7,9,13-15H,3H2,1H3,(H,11,16,17)/t5-,6-,7-,9-/m1/s1 |
| InChIKey | ZXIATBNUWJBBGT-JXOAFFINSA-N |
| Search the molecule in external databases | ChEMBL PubChem Compound Database Ligand Expo WIPO |
| PubChem CID | |
| PubChem SIDs |
9539104
24897322 37156244 76435034 113576922 129968611 135126211 135668174 162518632 223469912 226426237 250226135 252228288 252373654 252420358 252669156 254775008 256009543 274636362 310104005 310276420 312237548 319245459 321933889 329883574 341274327 342526814 349767373 355132892 363671175 374091534 375198021 375397473 377453065 377950786 382408487 387187402 388343173 404826212 419579744 438656714 439366901 439471142 440703506 443514087 |
* Chemical properties calculated with Open Babel - O'Boyle et al. Open Babel: An open chemical toolbox. J Cheminform 3, 33 (2011) (link)
QM Data:
| Dipole Magnitude [D]: | 5.985330215 |
| Energy [Eh]: | -1025.18181976419 |
| HOMO [eV]: | -9.2229 |
| LUMO [eV]: | 0.7021 |
| Gap [eV]: | 9.925 |
Download QM Data:
| Charges | charge.txt |
Download Structures
| 2D | .png .mol .mol2 .sdf .pdb .smi |
| 3D | .mol .mol2 .sdf .pdb |
Tautomers
| Tautomers SMILES |
COc1c(=O)[nH]c(=O)n(C2C(O)C(O)C(CO)O2)c1 tautomer #0
COc1c(O)nc(=O)n(C2C(O)C(O)C(CO)O2)c1 tautomer #1 COc1c(=O)nc(O)n(C2C(O)C(O)C(CO)O2)c1 tautomer #2 |
| Tautomer image | Show Image |
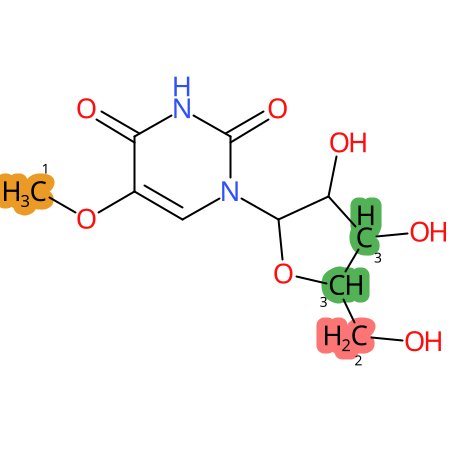
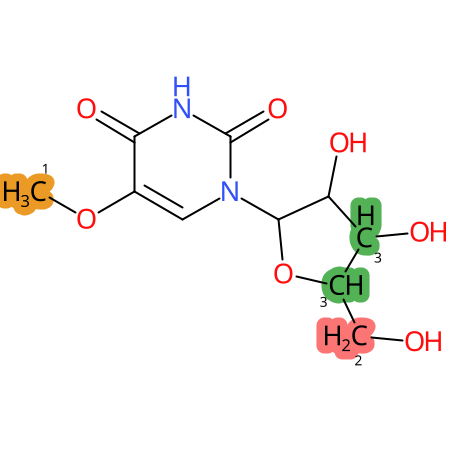
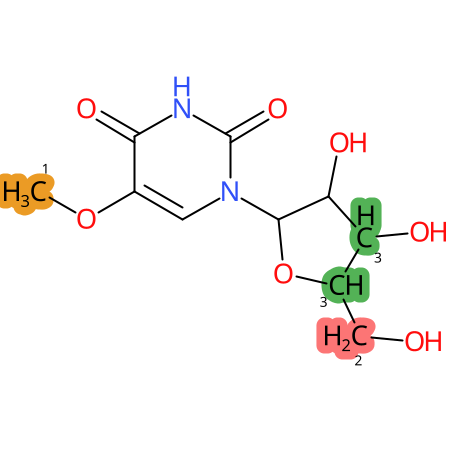
Predicted CYP Metabolic Sites
| CYP3A4 | CYP2D6 | CYP2C9 |
|---|---|---|
|
|
|
* CYP Metabolic sites predicted with SMARTCyp. SMARTCyp is a method for prediction of which sites in a molecule that are most liable to metabolism by Cytochrome P450. It has been shown to be applicable to metabolism by the isoforms 1A2, 2A6, 2B6, 2C8, 2C19, 2E1, and 3A4 (CYP3A4), and specific models for the isoform 2C9 (CYP2C9) and isoform 2D6 (CYP2D6). CYP3A4, CYP2D6, and CYP2C9 are the three of the most important enzymes in drug metabolism since they are involved in the metabolism of more than half of the drugs used today. The three top-ranked atoms are highlighted. See: SmartCYP and SmartCYP - background; Patrik Rydberg, David E. Gloriam, Lars Olsen, The SMARTCyp cytochrome P450 metabolism prediction server, Bioinformatics, Volume 26, Issue 23, 1 December 2010, Pages 2988–2989 (link)
LC-MS Information
| Monoisotopic mass | 274.0801 |
| Average mass | 274.227 |
| [M+H]+ | 275.0879 |
| Product ions | 143 |
| Normalized LC elution time * | not available |
| LC elution order/characteristics | not available |
* normalized to guanosine (G), measured with a RP C-18 column with acetonitrile/ammonium acetate as mobile phase.
Comments
mo5U is a modification characteristic for gram positive bacteria (B. subtilis, Firmicutes). Gram negative bacteria have cmo5U and mcmo5U in this position (E. coli, Protebacteria). Null mutation of cmoA in S. enterica results in the accummulation of tRNA containing mo5U34 and ho5U34.
Chemical groups contained
| Type | Subtype |
|---|---|
| other | methoxy |
Reactions producing 5-methoxyuridine
| Name |
|---|
| ho5U:mo5U |
Reactions starting from 5-methoxyuridine
| Name |
|---|
| mo5U:cmo5U |
Publications
| Title | Authors | Journal | Details | ||
|---|---|---|---|---|---|
| The modified wobble nucleoside uridine-5-oxyacetic acid in tRNAPro(cmo5UGG) promotes reading of all four proline codons in vivo. | Nasvall SJ, Chen P, Bjork GR | RNA | [details] | 15383682 | - |
| 5-methoxyuridine: a new minor constituent located in the first position of the anticodon of tRNAAla, tRNAThr, and tRNAVal from Bacillus subtilis. | Murao K, Hasegawa T, Ishikura H | Nucleic Acids Res | [details] | 825836 | - |
Last modification of this entry: Sept. 15, 2025