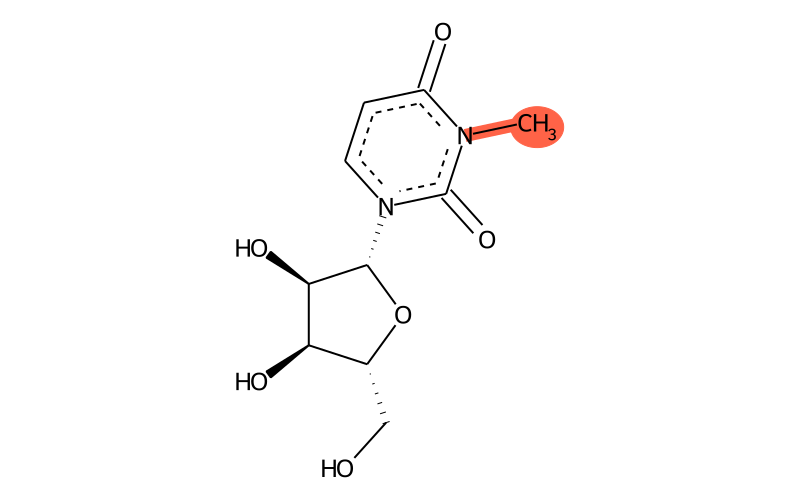
Summary
| Full name | 3-methyluridine |
| IUPAC name | 1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-3-methylpyrimidine-2,4-dione |
| Short name | m3U |
| MODOMICS code new | 2000000003U |
| MODOMICS code | 3U |
| Synonyms |
1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-3-methylpyrimidine-2,4-dione
1-((2R,3R,4S,5R)-3,4-Dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl)-3-methylpyrimidine-2,4(1H,3H)-dione 1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl]-3-methyl-pyrimidine-2,4-dione 2140-69-4 3-Methyluridine 86402-37-1 AC1L4106 AC1Q69A9 AR-1F4600 C008513 CID99592 CS-0061840 CTK1A5665 EX-A3664 HY-113138 N-3-Methyluridine N3-Methyluridine NSC 246077 SCHEMBL158123 Uridine, 3-methyl- Uridine, 3-methyl- (8CI)(9CI) UTQUILVPBZEHTK-ZOQUXTDFSA-N ZINC2558630 |
| Nature of the modified residue | Natural |
| RNAMods code | δ |
| Residue unique ID | 49 |
| Found in RNA | Yes |
| Related nucleotides | 199 |
| Enzymes |
Bmt5 (Saccharomyces cerevisiae) Bmt6 (Saccharomyces cerevisiae) RsmE (Escherichia coli) |
| Found in phylogeny | Eubacteria |
| Found naturally in RNA types | rRNA |
Chemical information
| Sum formula | C10H14N2O6 |
| Type of moiety | nucleoside |
| Degeneracy | not applicable |
| PubChem ID | 316991 |
| SMILES | C[n]1c(=O)[n]([C@@H]2O[C@H](CO)[C@@H](O)[C@H]2O)ccc1=O |
| logP | -2.8415 |
| TPSA | 113.92 |
| Number of atoms | 18 |
| Number of Hydrogen Bond Acceptors 1 (HBA1) | 6 |
| Number of Hydrogen Bond Acceptors 2 (HBA2) | 8 |
| Number of Hydrogen Bond Donors (HBD) | 3 |
| InChI | InChI=1S/C10H14N2O6/c1-11-6(14)2-3-12(10(11)17)9-8(16)7(15)5(4-13)18-9/h2-3,5,7-9,13,15-16H,4H2,1H3/t5-,7-,8-,9-/m1/s1 |
| InChIKey | UTQUILVPBZEHTK-ZOQUXTDFSA-N |
| Search the molecule in external databases | ChEMBL PubChem Compound Database Ligand Expo WIPO |
| PubChem CID | |
| PubChem SIDs |
10230690
15094871 44429785 57336922 79164892 103131464 104424131 117584450 129419024 135055823 140033790 143178712 162500138 172080103 226521886 249709617 250003447 252392747 256557955 273528947 304738260 341274389 342526820 349717701 363671424 374053368 375987068 375987319 378020676 383356576 385864089 388881998 401966905 405000900 415804679 433775050 433775447 433909743 434742392 438500018 439444366 |
* Chemical properties calculated with Open Babel - O'Boyle et al. Open Babel: An open chemical toolbox. J Cheminform 3, 33 (2011) (link)
QM Data:
| Dipole Magnitude [D]: | 6.224213292 |
| Energy [Eh]: | -950.011835703155 |
| HOMO [eV]: | -9.4272 |
| LUMO [eV]: | 0.886 |
| Gap [eV]: | 10.3132 |
Download QM Data:
| Charges | charge.txt |
Download Structures
| 2D | .png .mol .mol2 .sdf .pdb .smi |
| 3D | .mol .mol2 .sdf .pdb |
Tautomers
| Tautomers SMILES |
Cn1c(=O)n(C2OC(CO)C(O)C2O)ccc1=O tautomer #0
|
| Tautomer image | Show Image |
Predicted CYP Metabolic Sites
| CYP3A4 | CYP2D6 | CYP2C9 |
|---|---|---|
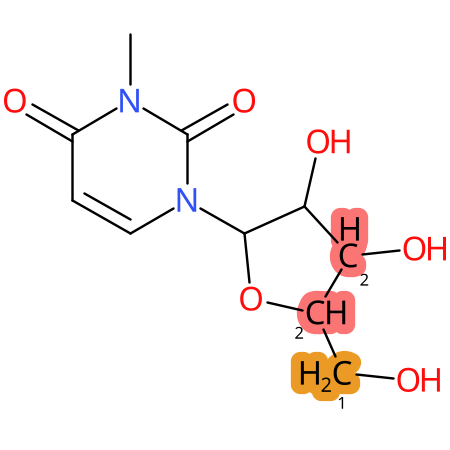
|
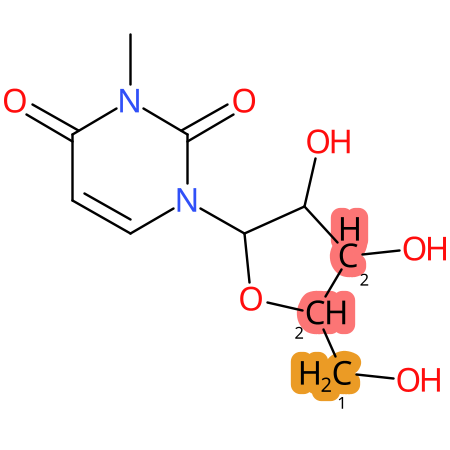
|
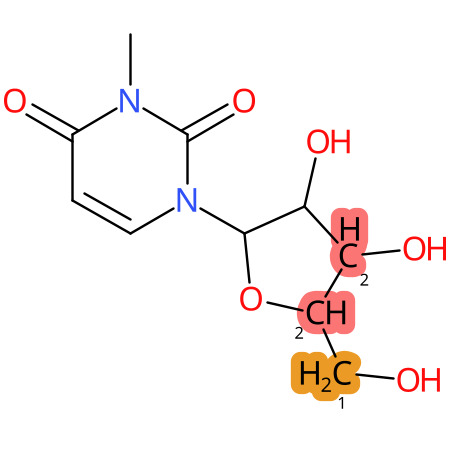
|
* CYP Metabolic sites predicted with SMARTCyp. SMARTCyp is a method for prediction of which sites in a molecule that are most liable to metabolism by Cytochrome P450. It has been shown to be applicable to metabolism by the isoforms 1A2, 2A6, 2B6, 2C8, 2C19, 2E1, and 3A4 (CYP3A4), and specific models for the isoform 2C9 (CYP2C9) and isoform 2D6 (CYP2D6). CYP3A4, CYP2D6, and CYP2C9 are the three of the most important enzymes in drug metabolism since they are involved in the metabolism of more than half of the drugs used today. The three top-ranked atoms are highlighted. See: SmartCYP and SmartCYP - background; Patrik Rydberg, David E. Gloriam, Lars Olsen, The SMARTCyp cytochrome P450 metabolism prediction server, Bioinformatics, Volume 26, Issue 23, 1 December 2010, Pages 2988–2989 (link)
LC-MS Information
| Monoisotopic mass | 258.0852 |
| Average mass | 258.228 |
| [M+H]+ | 259.093 |
| Product ions | 127 |
| Normalized LC elution time * | not available |
| LC elution order/characteristics | not available |
* normalized to guanosine (G), measured with a RP C-18 column with acetonitrile/ammonium acetate as mobile phase.
Chemical groups contained
| Type | Subtype |
|---|---|
| methyl group | methyl at aromatic O |
Reactions producing 3-methyluridine
| Name |
|---|
| U:m3U |
Reactions starting from 3-methyluridine
| Name |
|---|
| m3U:m3Um |
Last modification of this entry: Sept. 15, 2025