Since the advent of DNAzymes, these catalysts have found application in a variety of research and laboratory activities. The most attractive aspect of utilizing DNAzymes is the possibility to have them “tailor-made” to our needs, that is, depending on the precise substrates we want them to act upon.
DNAzymeBuilder relies on a database that contains information on 44-RNA and 93-DNA cleaving DNAzymes, including the reaction conditions under which they best operate, their kinetic parameters, the type of cleavage reaction that is catalyzed, the specific sequence that is recognized by the DNAzyme, the cleavage site within this sequence, and special design features that might be necessary for optimal activity of the DNAzyme. Based on this information and the input sequence provided by the user, DNAzymeBuilder is able to provide a list of DNAzymes to carry out the cleavage reaction and detailed information for each of them, including the expected yield, reaction products and optimal reaction conditions.
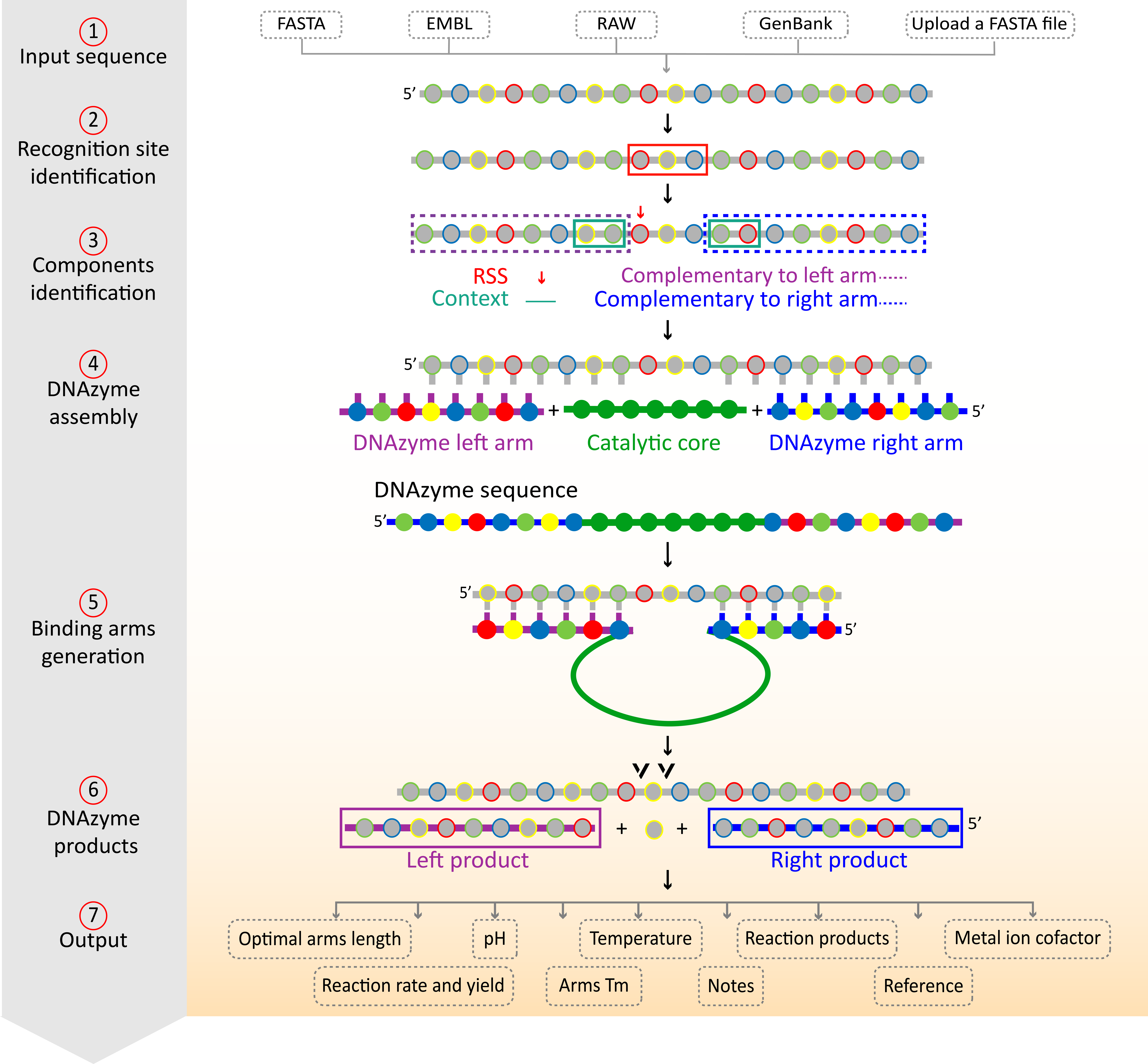
What DNAzymeBuilder does:
1- It examines the query for k-mers (a short specific set of nucleotides) that match the preferred cleaving sites of the DNAzymes present in the database (recognition site). Since the nucleotides adjacent to the recognition site, as well as the cleavage site itself, may influence the cleavage yield, the predicted efficiency of catalysis is reported for each case in the notes section of the output.
2- It provides optimal reaction conditions for the chosen DNAzymes, including temperature, pH, arm lengths, and required cofactors. The reported reaction rate is mentioned for each set of reaction conditions.
3- It reports the type of cleavage reaction. The functional groups at 5’ and 3’ ends of the products are presented when data were available in the literature.
4- It assembles DNAzymes with binding arms that may or may not be fully base-paired to the substrates. While DNAzymes’ binding arms are often fully base-paired to the substrates, in some cases bulges or unpaired nucleotides are required for maximal catalytic activity. The appropriate information will appear in the notes section of the output.
F-8
10-23
Sequence elements in a DNAzyme-substrate complex recognized by DNAzymeBuilder
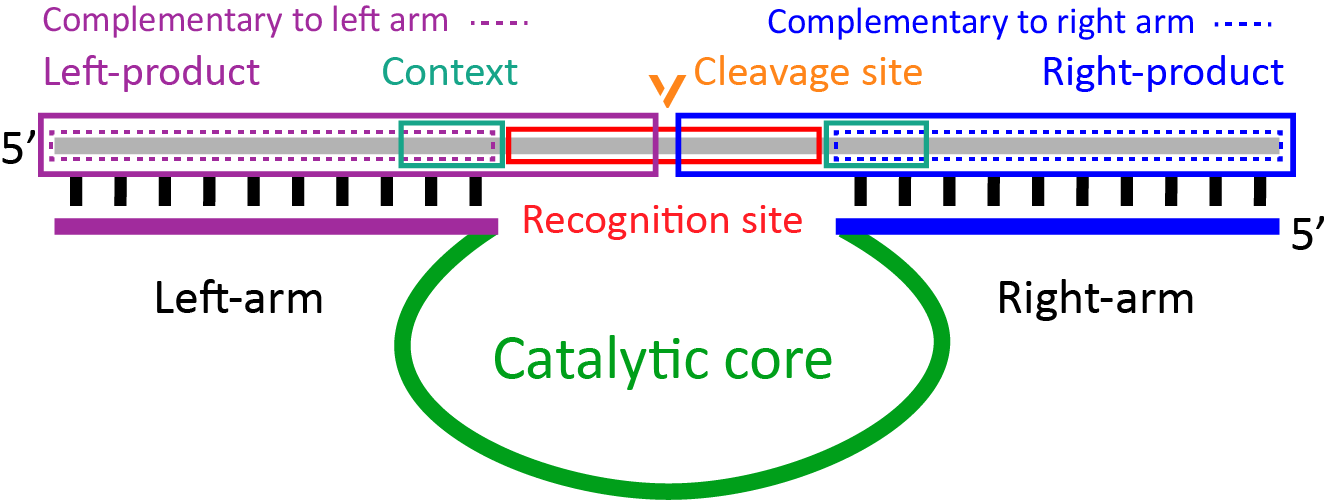
DNAzymeBuilder operates taking into account the following sequence elements (Figure 1):
1- Cleavage site (CS): A cleavage site represents the exact position of the phosphodiester bond that is cleaved. For DNAzymes that only cleave once, the phosphodiester bond is defined by specifying the flanking nucleotides. For DNAzymes that cleave more than once, the excised nucleotides are marked as the cleavage site.
2- Recognition site (RS): A recognition site designates the required sequence on the substrate that is recognized by the DNAzyme. It may encompass the cleavage site, however, there are cleaving DNAzymes for which the cleavage site is not within the RS. Most DNAzymes have only one RS but some have more.
3- Recognition site start (RSS): It is the starting point of the recognition site. The first nucleotide in the recognition sequence is marked as RSS and it serves as the starting point to calculate the beginning of the DNAzyme’s binding arms.
4- Left and right binding arms: The binding arms of the DNAzyme are the sequences to the left and right of the catalytic core acting as scaffolds for the substrate. They are designed based on the RSS point and their optimal lengths are chosen based on the data reported in the original articles. Nonetheless, users may specify the binding arms’ length, or select it based on the desired Tm.
5- Catalytic core: The catalytic core sequence is specific to each DNAzyme. However, some DNAzymes may still be catalytically active in mutant forms. If point-mutants or a given mutation in the catalytic core are known to lead to an increased catalytic activity of the DNAzyme, the output of DNAzymeBuilder includes both the originally reported and the mutant catalytic cores.
6- Context: The nucleotides upstream and downstream from the RS constitute the context, that affects the activity of the DNAzyme.
Construction of the cleaving DNAzymes
After the user provides the (target) substrate sequence, the search for the recognition site begins. If there is a recognition site, i.e. the substrate is cleavable by at least one of the DNAzymes in the database, DNAzymeBuilder generates the binding arms for the DNAzyme(s) that will cleave this substrate. The right and left arms of the DNAzyme are made to be complementary to the substrate and are written in reverse (5’ to 3’). DNAzymeBuilder can assemble DNAzymes in three different ways:
1. Based on optimal arm length: In this procedure, the lengths of the arms are set based on the optimal lengths reported in the literature. If no such lengths have been reported, a default value (10 nucleotides) is used.
2. Based on the user's specifications: users may specify the lengths of the right and left arms of the DNAzyme. In this case, the portion of the substrate sequence that binds/hybridizes to the DNAzyme depends on the user's specified arm lengths.
3. Based on the desired Tm: users may specify the Tm for the right and left arm duplexes. In this case, the lengths of the binding arms are determined by an algorithm that calculates the Tm of binding arms with increasing lengths. In other words, based on the given sequence, the algorithm specifies how many nucleotides are required at the right and left binding arms to allow for Tm value that is equal or just above the input Tm.
The final step consists on assembling the catalytic core and the right and left binding arms. The resulting DNAzyme sequence is presented 5’ to 3’.