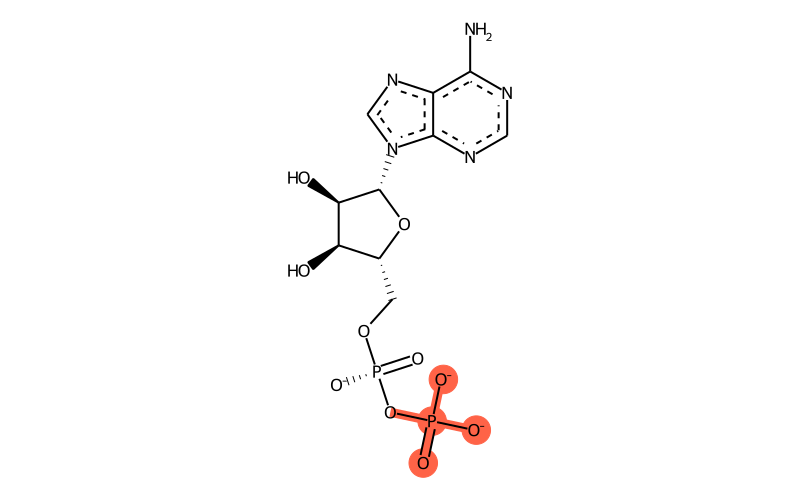
Summary
| Full name | adenosine-5'-diphosphate |
| IUPAC name | [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy-oxidophosphoryl] phosphate |
| Short name | ppA |
| MODOMICS code new | 2000000552A |
| MODOMICS code | 552A |
| Synonyms |
20398-34-9
2e2p 2fr8 2hmv 2ja3 2o1v 2qv7 [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanidyl-phosphoryl] phosphate [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-2-oxolanyl]methoxy-oxidophosphoryl] phosphate [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy-oxidophosphoryl] phosphate 2r6f 2rio 2vhj 2y8i 2y8l 2y8q 2z4r 2zfi 2zkj 3a1d 3atu 3atv 3ay9 3bf1 3bxz 3c7n 3cf1 3d2r 3dgn 3i73 3k1j 3llm 3mle 3ncr 3p23 3qc9 3qxh 3rup 3rv3 3rv4 3t4n 3ta1 3tif 4i3z 58-64-0 5'-O-[(phosphonatooxy)phosphinato]adenosine 9-{5-O-[(Phosphonatooxy)phosphinato]pentofuranosyl}-9H-purin-6-amine A831962 AC1OEJTD Adenosine 5'-diphosphate adenosine 5'-pyrophosphate adenosine pyrophosphate adenosine-5-diphosphate adenosine-5'-diphosphate adenosine-diphosphate ADP ADP trianion ADP(3-) CHEBI:456216 CID7058055 DTXSID70942564 Q27225748 ZINC03861153 |
| Nature of the modified residue | Natural |
| RNAMods code | か |
| Residue unique ID | 317 |
| Found in RNA | Yes |
| Related nucleosides | 109 |
Chemical information
| Sum formula | C10H12N5O10P2 |
| Type of moiety | nucleotide |
| Degeneracy | not aplicable |
| SMILES | [O-][P@@](OC[C@H]1O[C@H]([C@H](O)[C@@H]1O)[n]1c[n]c2c1[n]c[n]c2N)(OP([O-])(=O)[O-])=O |
| logP | 0.1498 |
| TPSA | 260.71 |
| Number of atoms | 27 |
| Number of Hydrogen Bond Acceptors 1 (HBA1) | 11 |
| Number of Hydrogen Bond Acceptors 2 (HBA2) | 15 |
| Number of Hydrogen Bond Donors (HBD) | 3 |
| PDB exact match , link to the most similar ligand | ADP |
| InChI | InChI=1S/C10H15N5O10P2/c11-8-5-9(13-2-12-8)15(3-14-5)10-7(17)6(16)4(24-10)1-23-27(21,22)25-26(18,19)20/h2-4,6-7,10,16-17H,1H2,(H,21,22)(H2,11,12,13)(H2,18,19,20)/p-3/t4-,6-,7-,10-/m1/s1 |
| InChIKey | XTWYTFMLZFPYCI-KQYNXXCUSA-K |
| Search the molecule in external databases | ChEMBL ChemAgora ChEBI PubChem Compound Database Ligand Expo ChemSpider WIPO |
| PubChem CID | |
| PubChem SIDs |
2115
17201474 43647309 50737653 53812622 53812680 53812741 85646635 87557157 87557308 87557345 87557366 87557477 87557537 87557593 87557619 114846720 131319866 170481166 170481337 170481801 170481802 170481804 170481915 170481927 170481966 170482031 170482032 170482053 170482096 170482144 170482178 170482785 170482918 170483197 170483292 170483403 170483609 170483821 170483909 170484002 170484004 170484005 170484096 170484257 170484291 170485158 384499616 385730366 386285270 386566985 427204810 |
* Chemical properties calculated with Open Babel - O'Boyle et al. Open Babel: An open chemical toolbox. J Cheminform 3, 33 (2011) (link)
Download Structures
| 2D | .png .mol .mol2 .sdf .pdb .smi |
| 3D | .mol .mol2 .sdf .pdb |
Tautomers
| Tautomers SMILES |
[O-]P(OCC1OC(C(O)C1O)n2cnc3c2ncnc3N)(OP([O-])(=O)[O-])=O tautomer #0
[O-]P(OCC1OC(C(O)C1O)n2cnc3c2ncnc3N)(OP([O-])(=O)[O-])=O tautomer #1 [O-]P(OCC1OC(C(O)C1O)n2cnc3c2[nH]cnc3=N)(OP([O-])(=O)[O-])=O tautomer #2 [O-]P(OCC1OC(C(O)C1O)n2cnc3c2nc[nH]c3=N)(OP([O-])(=O)[O-])=O tautomer #3 [O-]P(OCC1OC(C(O)C1O)N2C=NC3C2=NC=NC3=N)(OP([O-])(=O)[O-])=O tautomer #4 |
| Tautomer image | Show Image |
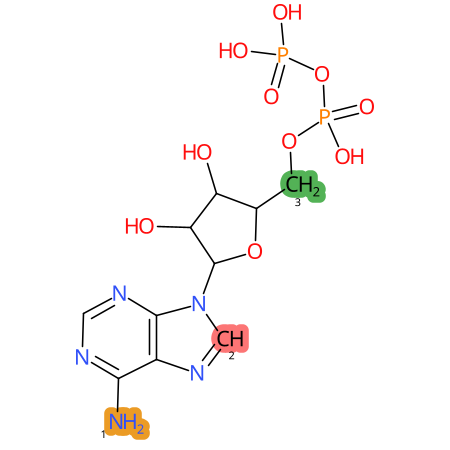
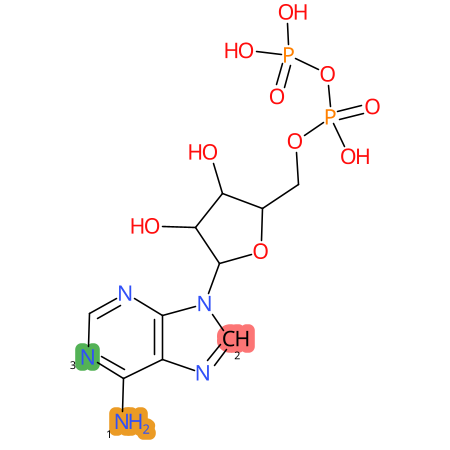
Predicted CYP Metabolic Sites
| CYP3A4 | CYP2D6 | CYP2C9 |
|---|---|---|
|

|
|
* CYP Metabolic sites predicted with SMARTCyp. SMARTCyp is a method for prediction of which sites in a molecule that are most liable to metabolism by Cytochrome P450. It has been shown to be applicable to metabolism by the isoforms 1A2, 2A6, 2B6, 2C8, 2C19, 2E1, and 3A4 (CYP3A4), and specific models for the isoform 2C9 (CYP2C9) and isoform 2D6 (CYP2D6). CYP3A4, CYP2D6, and CYP2C9 are the three of the most important enzymes in drug metabolism since they are involved in the metabolism of more than half of the drugs used today. The three top-ranked atoms are highlighted. See: SmartCYP and SmartCYP - background; Patrik Rydberg, David E. Gloriam, Lars Olsen, The SMARTCyp cytochrome P450 metabolism prediction server, Bioinformatics, Volume 26, Issue 23, 1 December 2010, Pages 2988–2989 (link)
LC-MS Information
| Monoisotopic mass | None |
| Average mass | 424.177 |
| [M+H]+ | not available |
| Product ions | not available |
| Normalized LC elution time * | not available |
| LC elution order/characteristics | not available |
* normalized to guanosine (G), measured with a RP C-18 column with acetonitrile/ammonium acetate as mobile phase.
Last modification of this entry: Sept. 22, 2023