GDFuzz3D: protein 2D map to 3D structure retrieval service 
Overview
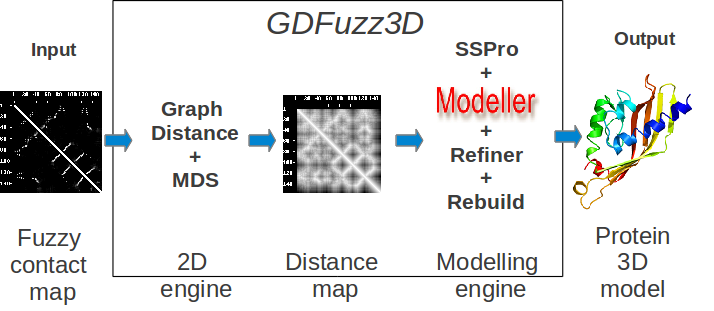
GDFuzz3D is a new method for protein tertiary structure retrieval from a contact map. It can handle binary (i.e. native) contact maps but the algorithm is designed to have predicted contact maps on input. The said map should be in CASP RR format, one of the formats supported by PROTMAP2D program as its input/output. The program can be also used for viewing and manipulating predicted (fuzzy) contact maps as well as calculating contact maps out of 3D models (i.e. GDFuzz3D final model).
An example of a fuzzy contact map in CASP RR format is here which gives such 3D output (visualized by PyMOL). Detailed CASP RR specification is to be found in the Appendix A of this pdf document or in the help section. Input map should contain the amino acid sequence as its SEQRES entries (one-letter code). It is also possible to input the amino acid sequence as a block of rows of maximum length of 50 aa. If no CASP RR 2D map is available but instead, one of matrix format, please refer to the Transform matrix tool for conversion into CASP RR.
GDFuzz3D makes use of a new non-Euclidean distance function as explained in the publication (presently submitted). Then it uses Multi-Dimensional Scaling algorithm to produce a crude 3D model. Subsequently, other bioinformatics programs are used in order to refine the model: SSPro, Modeller, Refiner, MMTSB Rebuild. The results are sent by email as a PDB file containing the predicted 3D model, based on the given fuzzy contact map. The approximate distance map produced before the 3D modelling procedure is started is attached in the email as well.
Contact: Michal J. Pietal,
Lukasz P. Kozlowski, Janusz M. Bujnicki,
Laboratory of Bioinformatics and Protein Engineering, International Institute of Molecular and Cell Biology in Warsaw