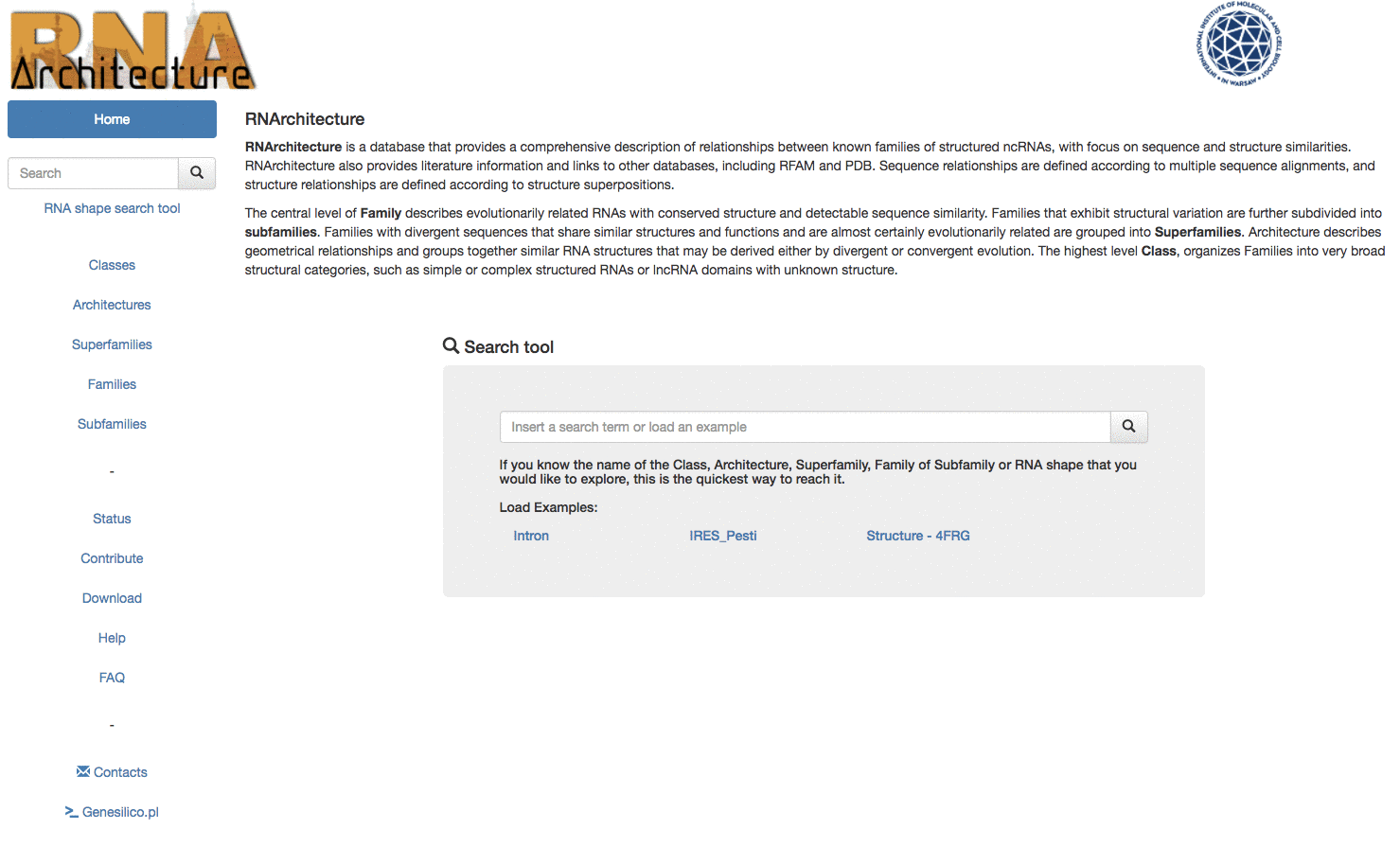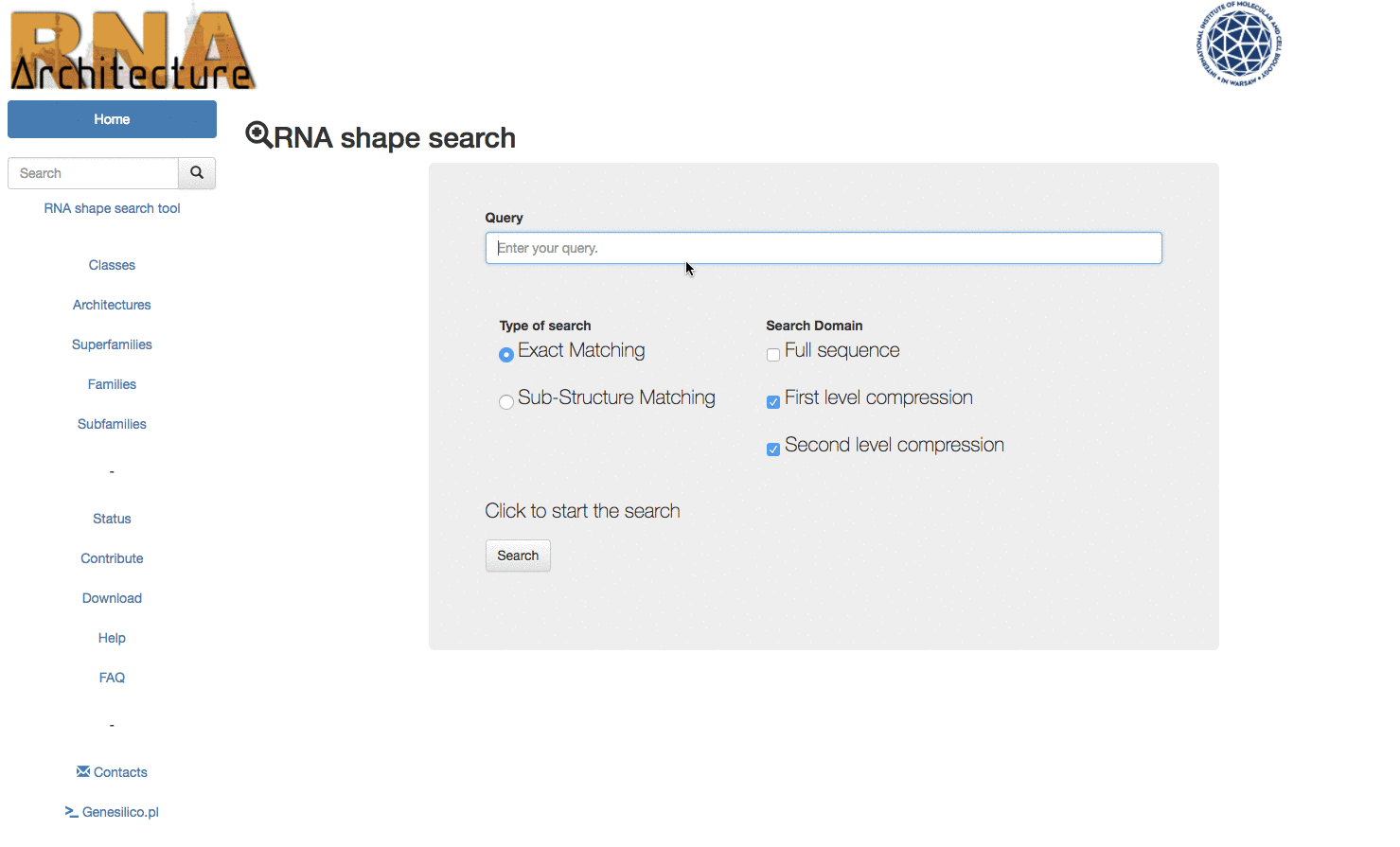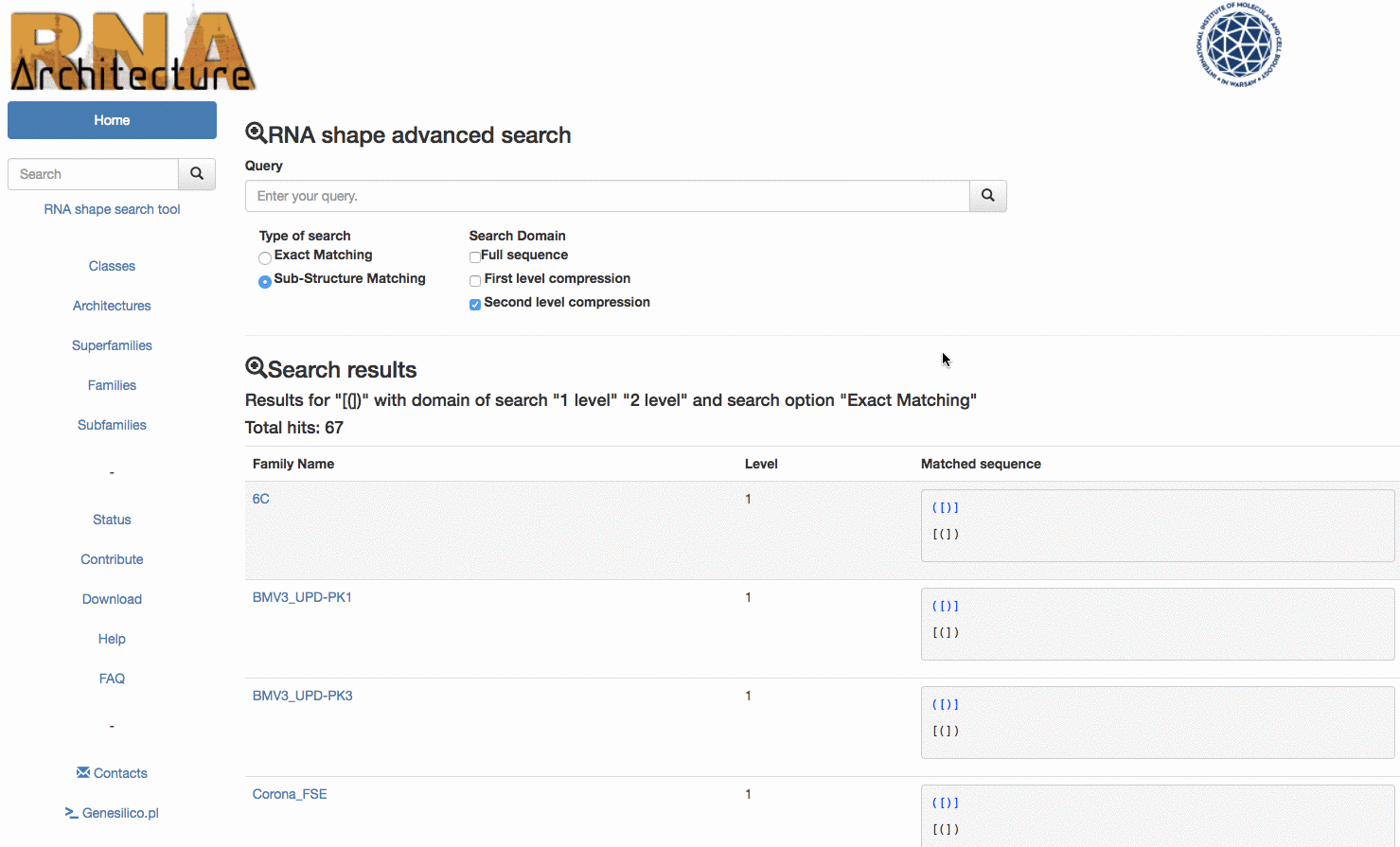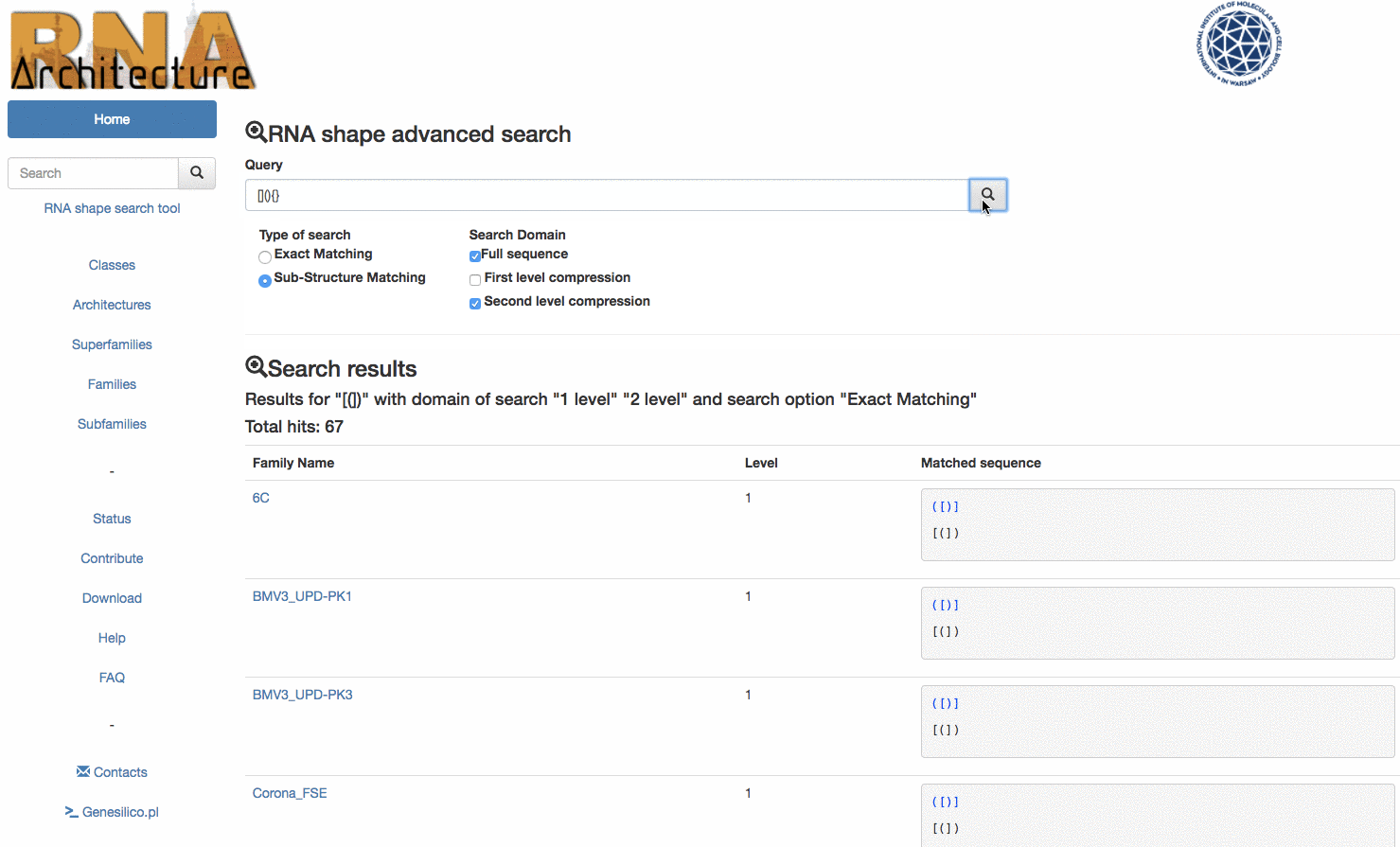
The RNArchitecture database has been developed to provide a comprehensive classification system that describes relationships between RNA families, with a focus on structural similarities.
| Browse Browse through the hierarchy. |
Search Search an entry or a subset in the database. |
RNA shape search Search a Family by its RNA shape |
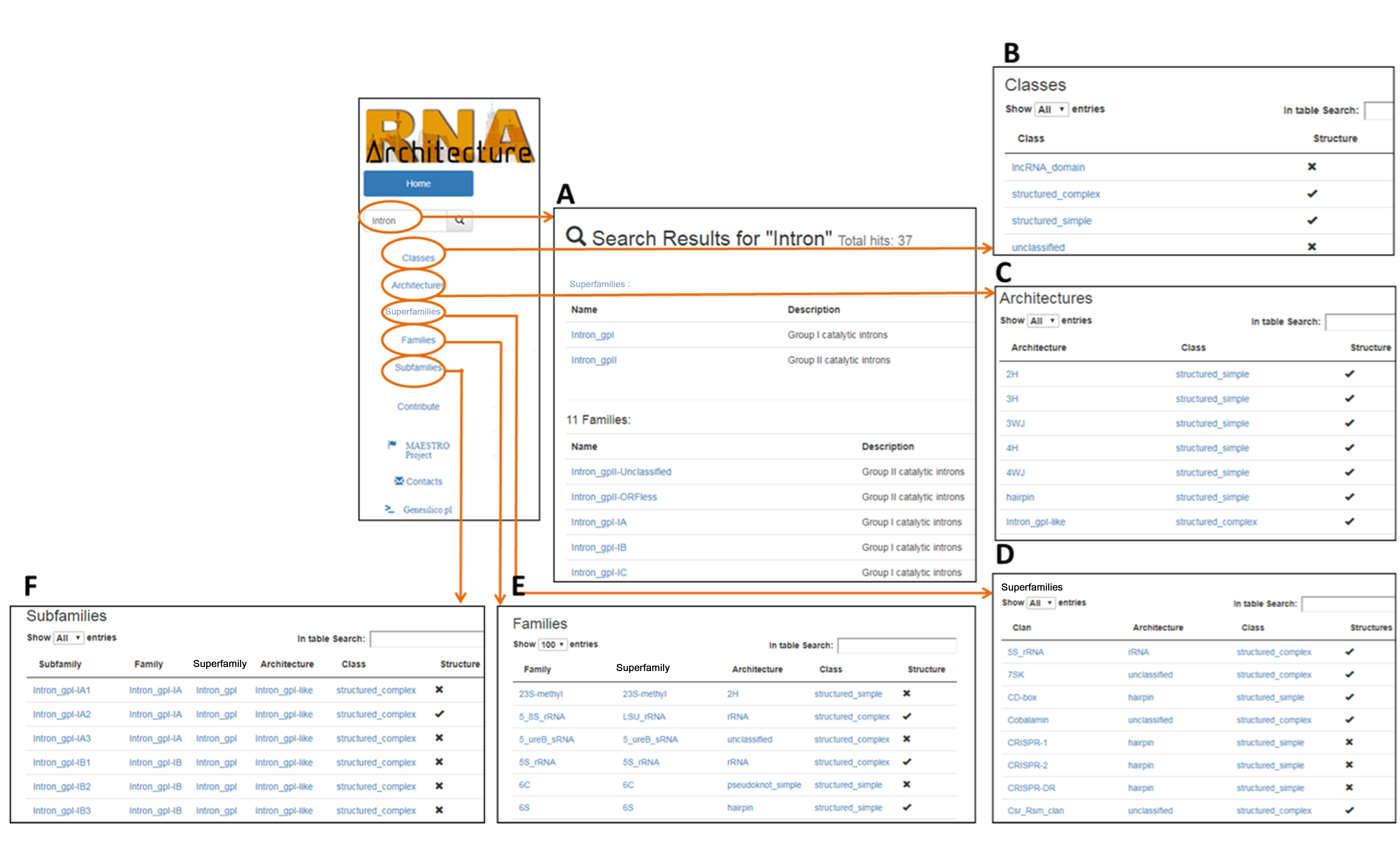
Fig.1 - Screenshots of RNArchitecture database. (A) Search result page, (B)Classes classification, (C) Architectures classification, (D) Superfamilies classification, (E) Families classification, (F) Subfamilies classification.
The central level of the classification is Family. Families group together evolutionarily related RNAs with conserved structure and detectable sequence similarity.
Families whose members exhibit structural variation are further subdivided into Subfamilies.
Families with similar structures and functions, and likely to be evolutionarily related are grouped into Superfamilies.
Families that share a similar core structure, but which are not clear homologs, are grouped into Architectures.The highest level, Class, organizes the data into very broad structural and functional categories.
The information of the database can be browsed using the left menu panel (Fig.1 - B,C,D,E,F). From here you can access any level of the classification. While accessing one of the levels above family, like Classes( Fig. 1 - B ) allows to surf the classification, reducing the number of displayed subsequent sub-levels, entering the classification in any other point, (es. Architectures) let the user have a list of all architectures available in the database.
In the following animations you have an example on how the database is browsed.
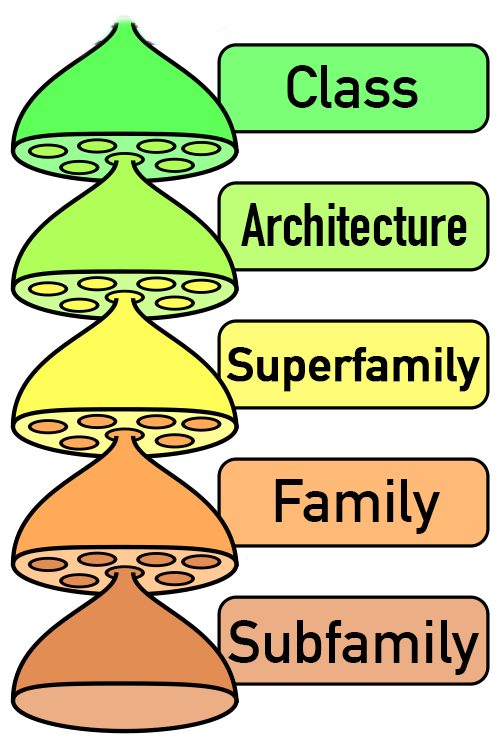 Fig. 2 - RNArchitecture hierarchical system
Fig. 2 - RNArchitecture hierarchical system
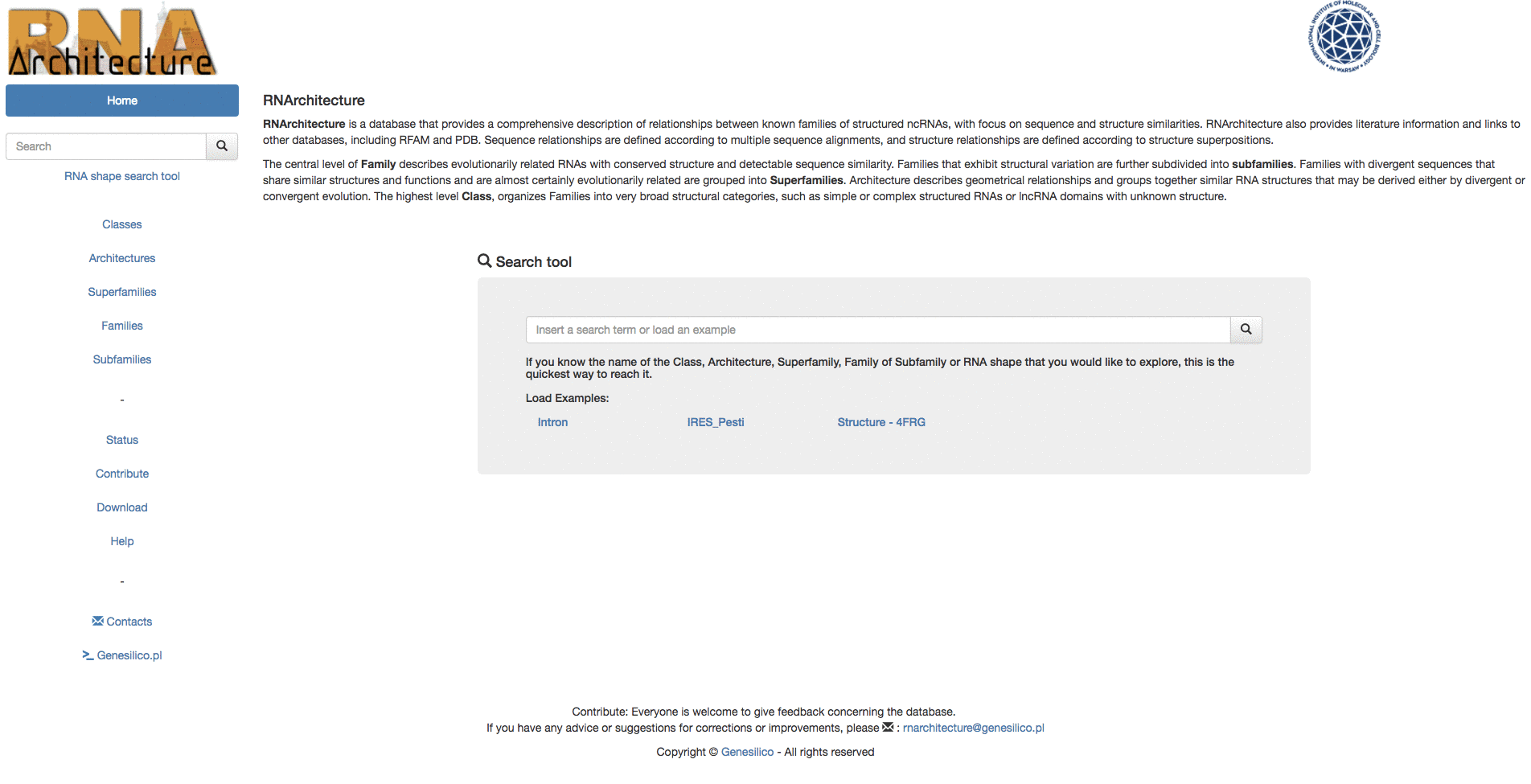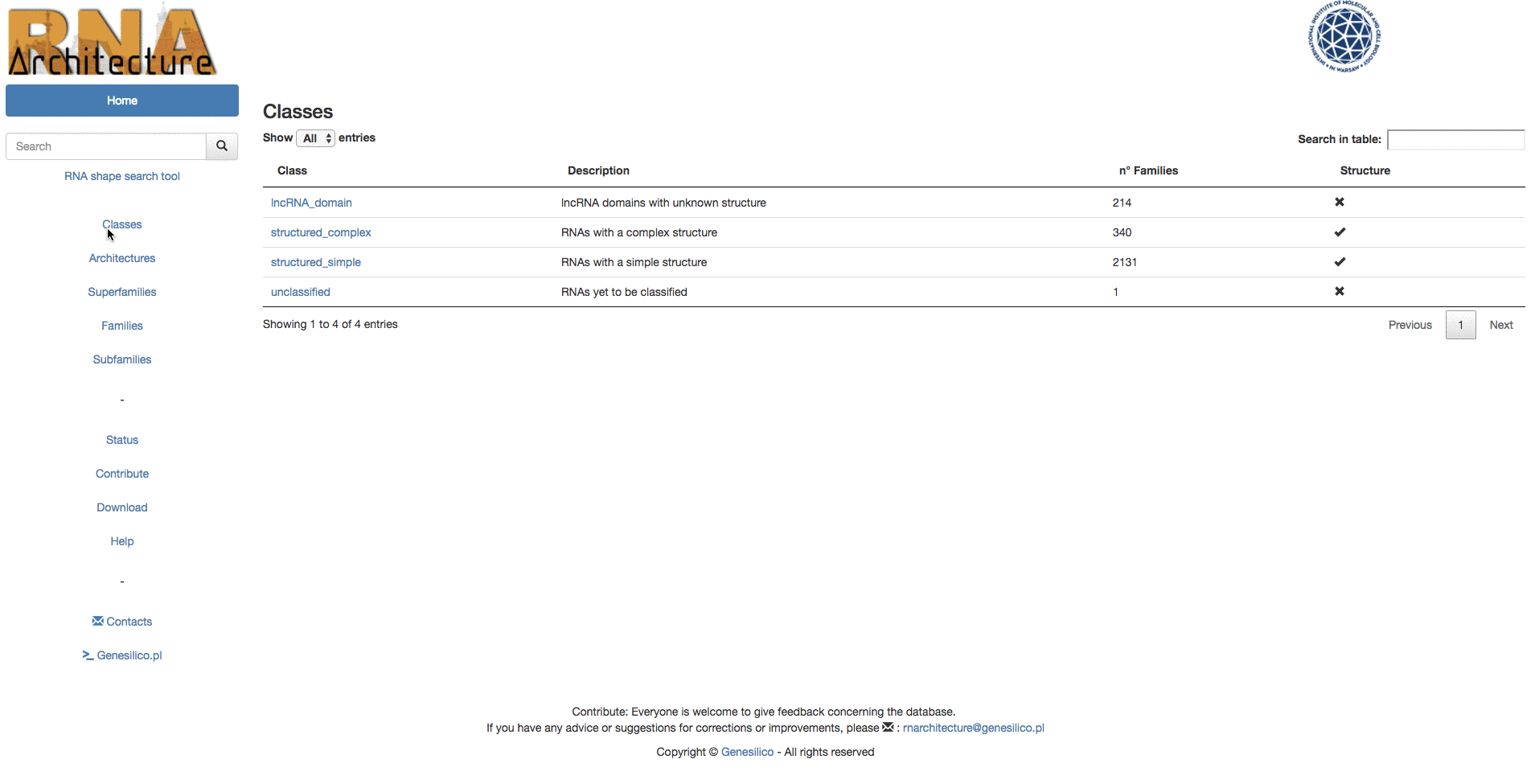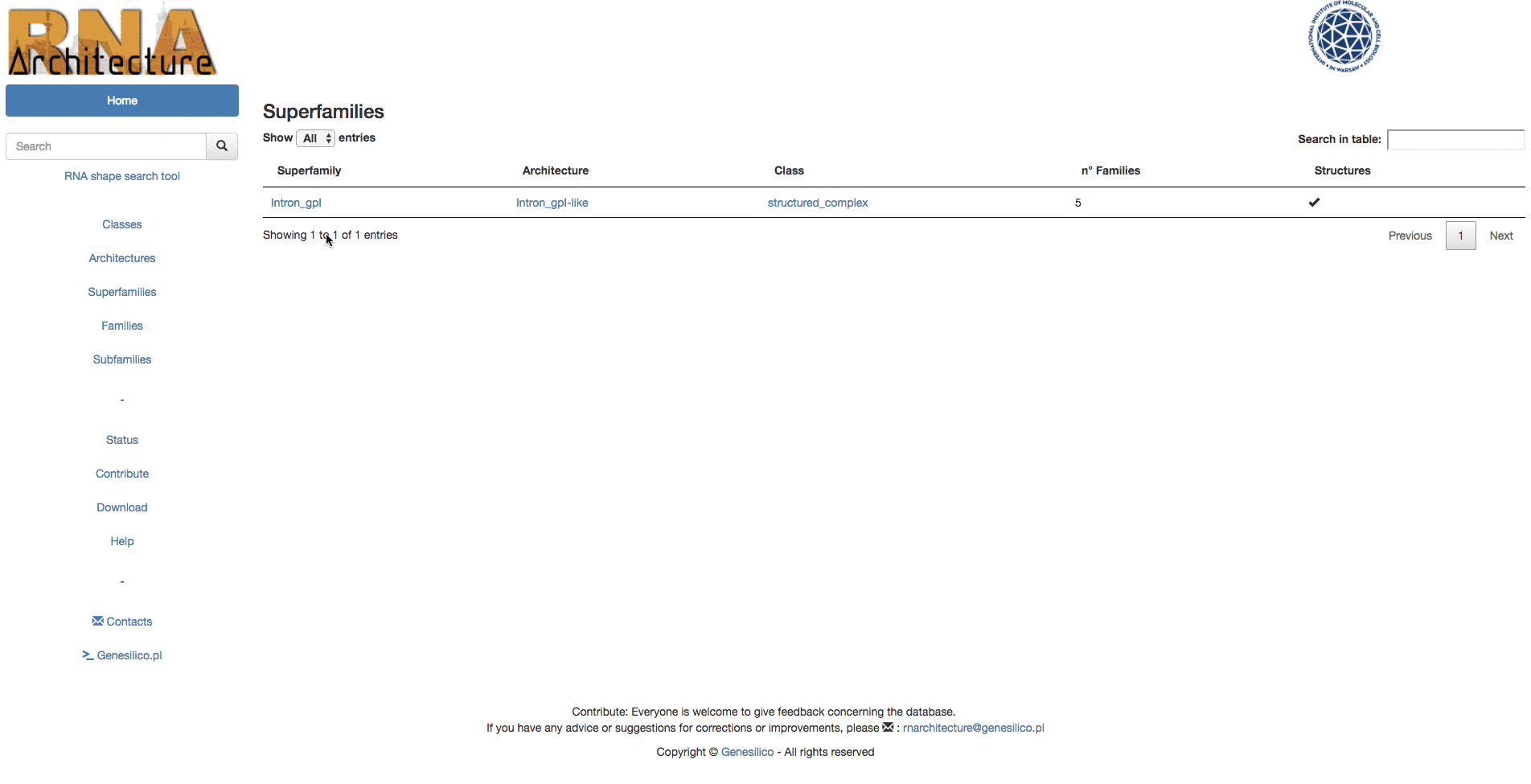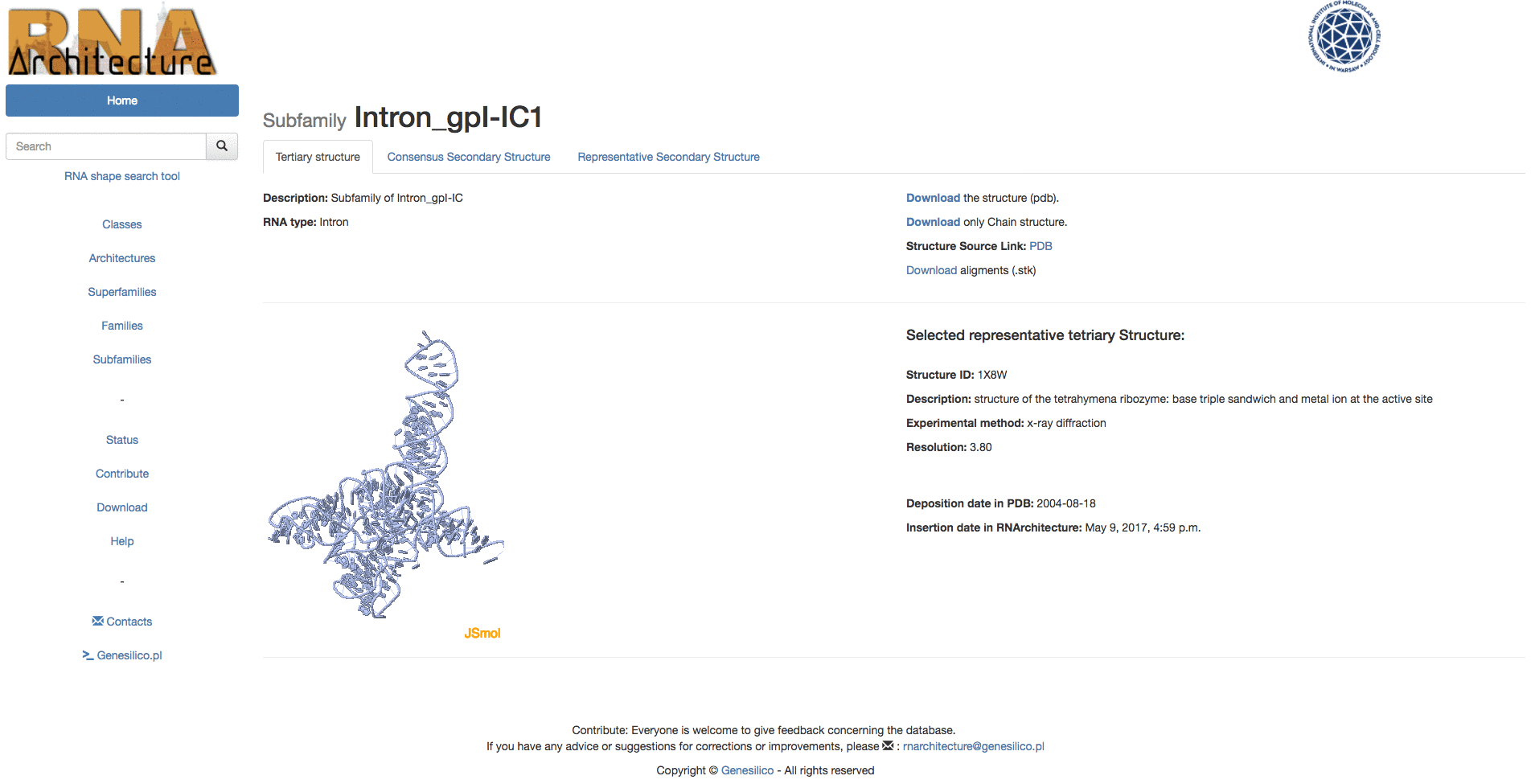 Example: Browsing the database from the top of the hierarchy to the subfamily "Intron_gpI_IC1".
Example: Browsing the database from the top of the hierarchy to the subfamily "Intron_gpI_IC1".
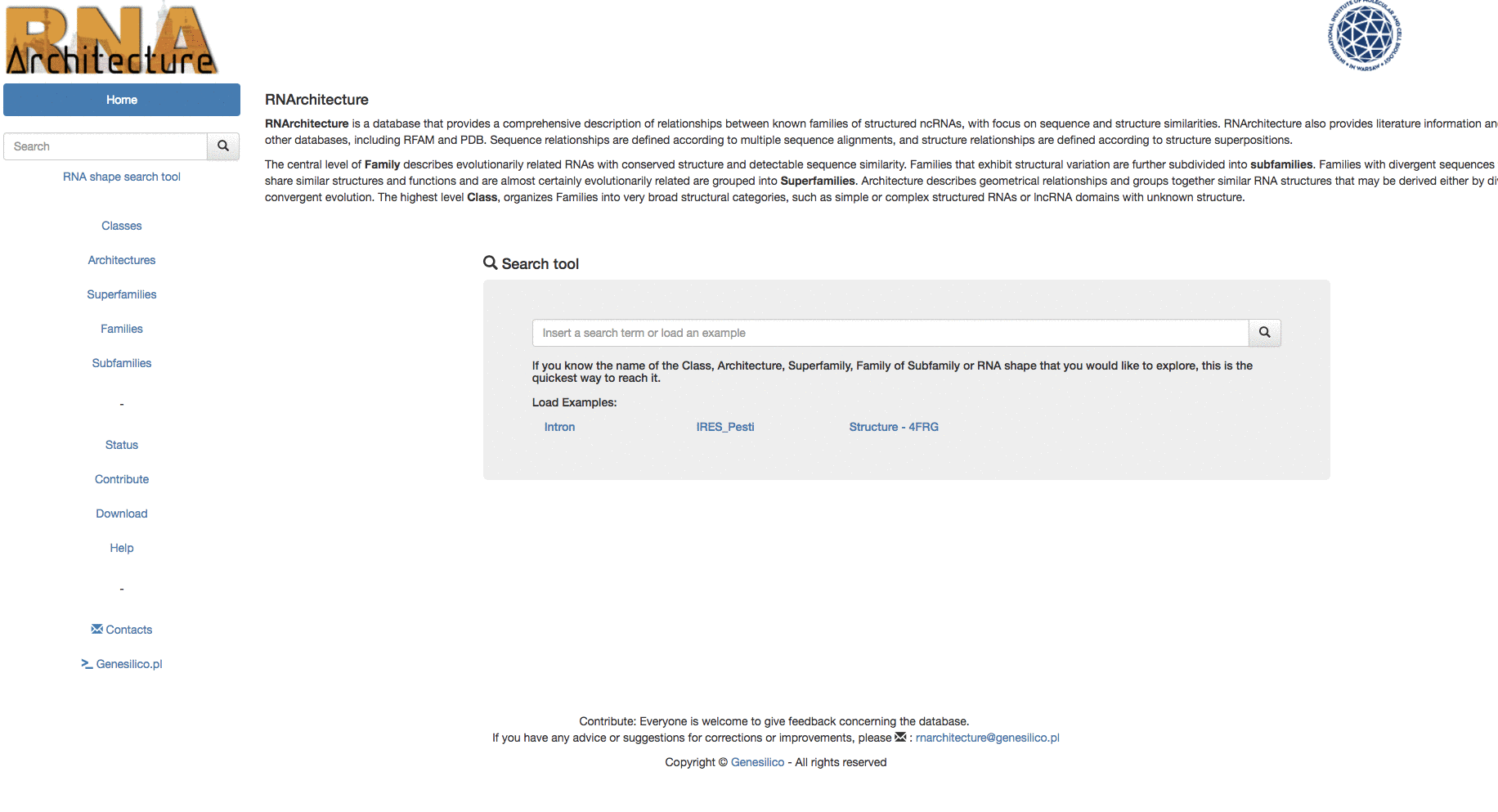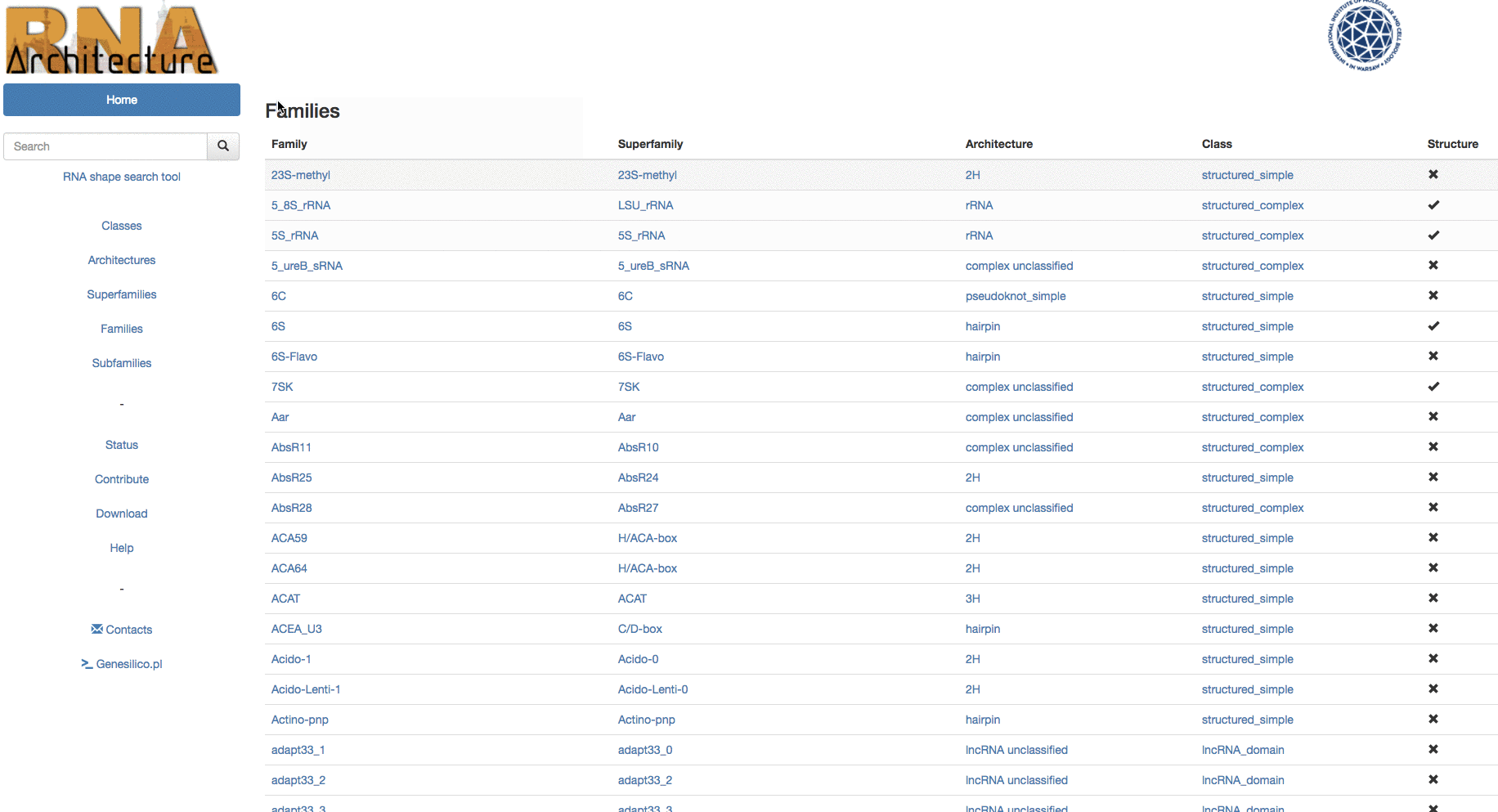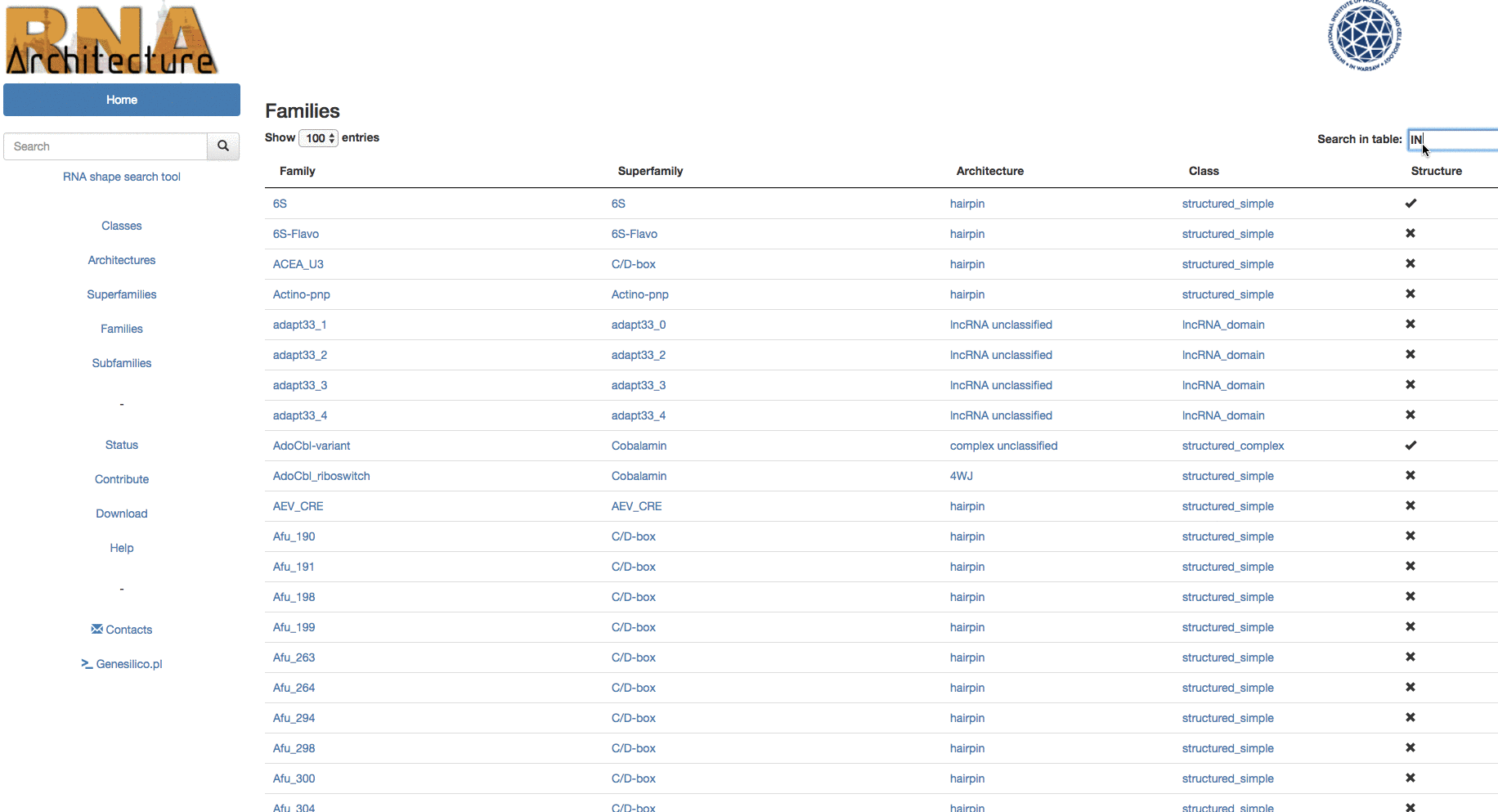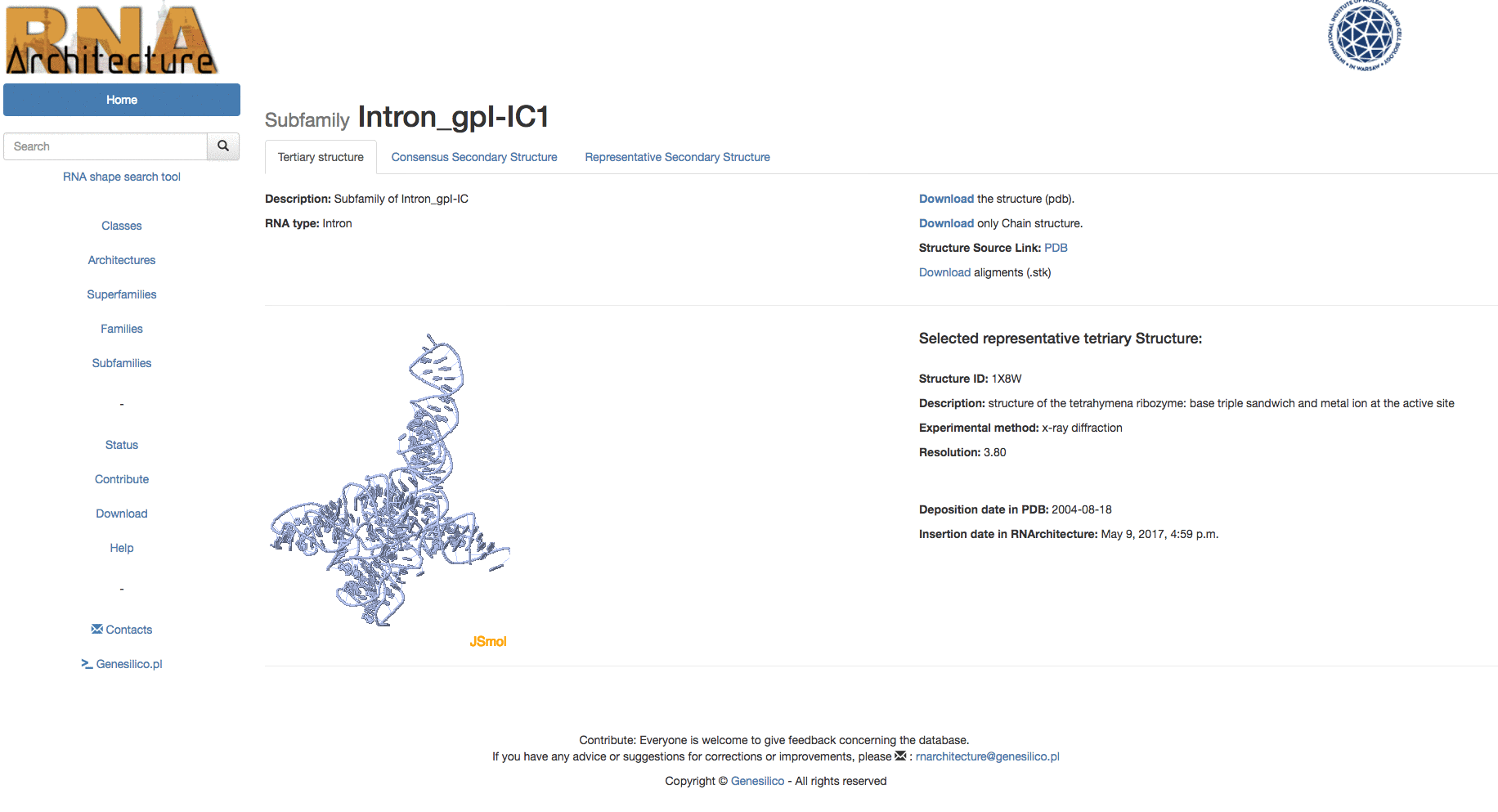 Example: Browsing from families, in table search in families to the subfamily "Intron_gpI_IC1".
Example: Browsing from families, in table search in families to the subfamily "Intron_gpI_IC1".
Options for database searching and querying have been implemented (Fig.1 - A), including search using:
| Search options | Examples |
|---|---|
| PDB IDS | 4FRG |
| Class | structured simple |
| Architecture | 4H |
| Superfamily | 6S-Flavo |
| Family | 5 8S rRNA |
| Subfamily | Intron gpI |
| Rfam accession number | RF01848 |
| RNA type | riboswitch |
The basic search can be performed from the home page of the website ( Fig.3 - 1 ) or from the omnipresent left menu bar ( Fig.3 - 2 ).
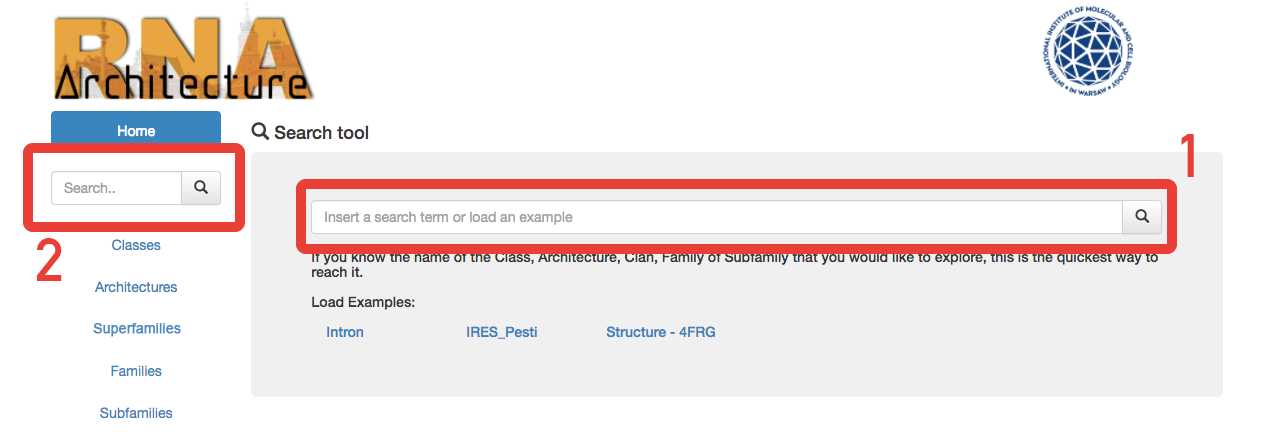 Fig. 3 - The search can be performed from the area (1) and (2) of the homepage.
Fig. 3 - The search can be performed from the area (1) and (2) of the homepage.
In the following animations you have an example on how to perform a search query on the database.
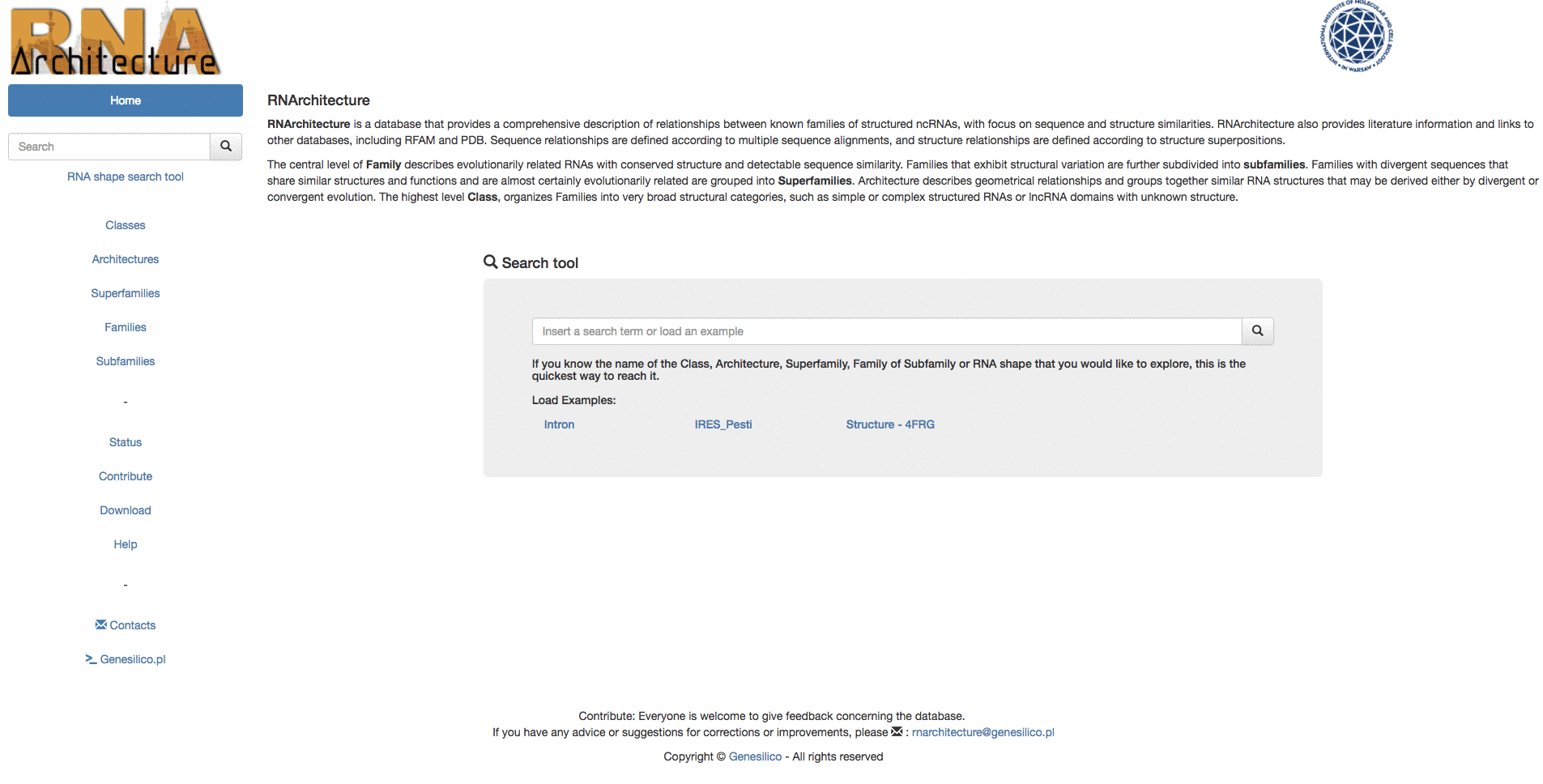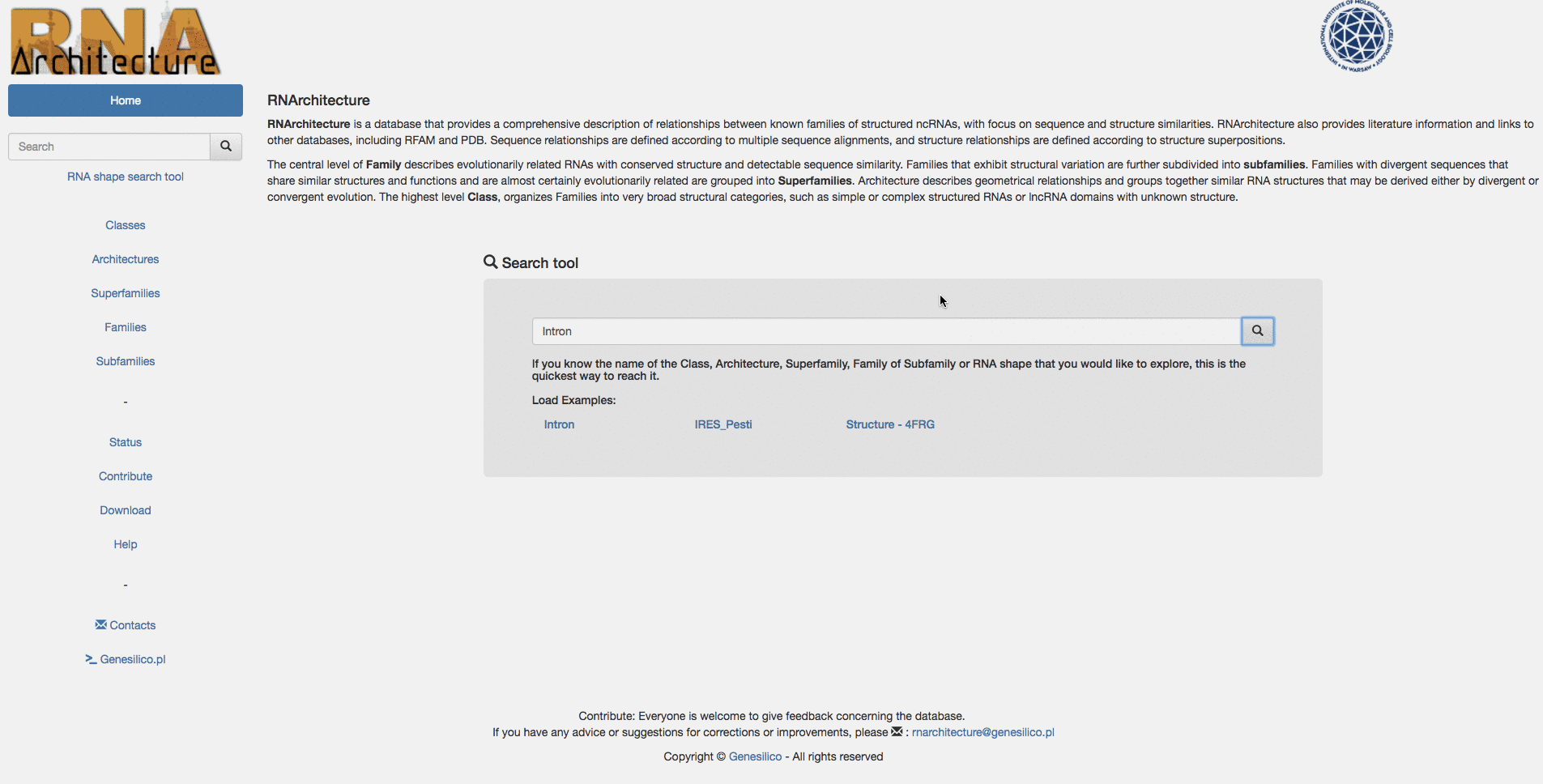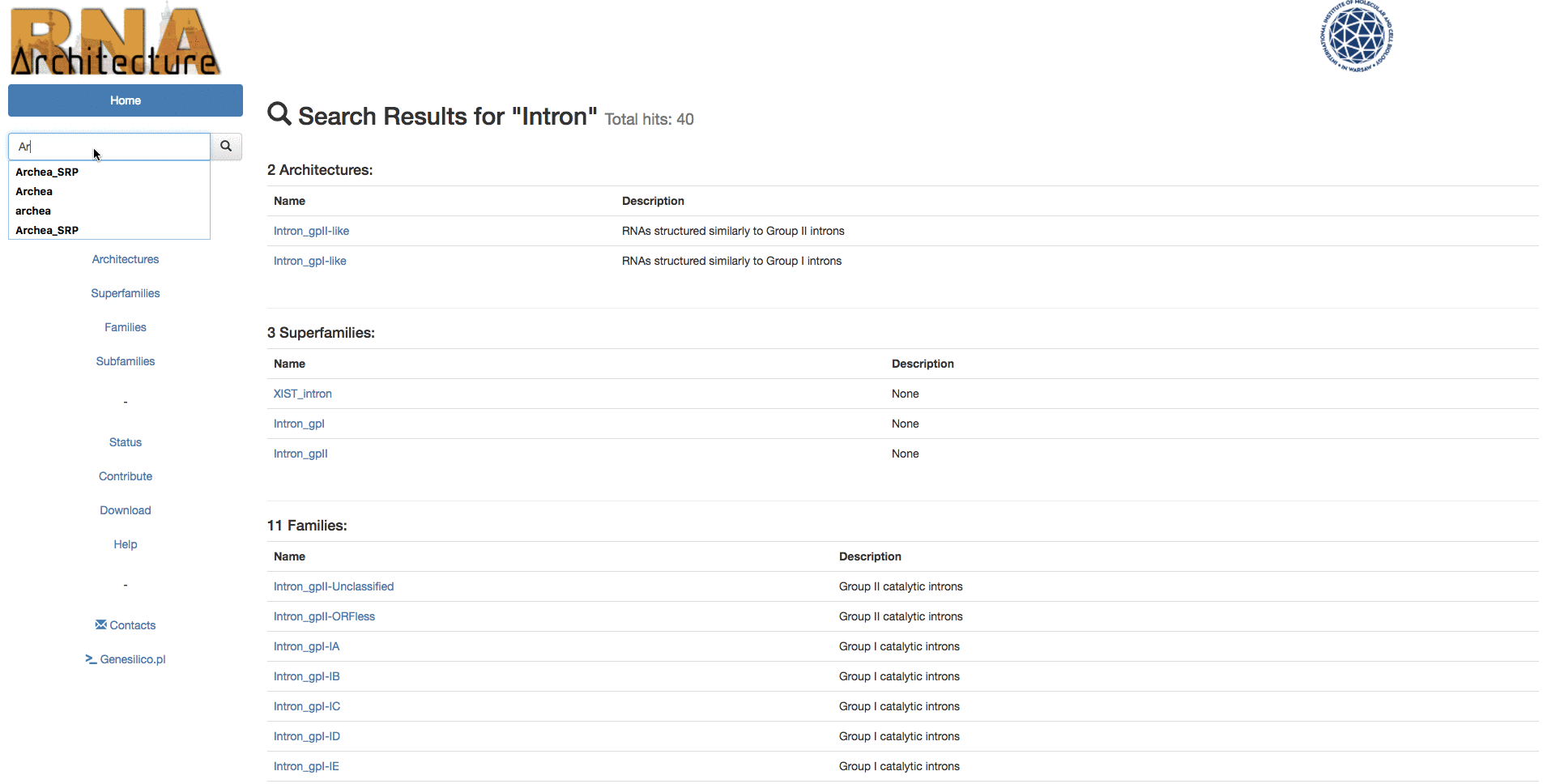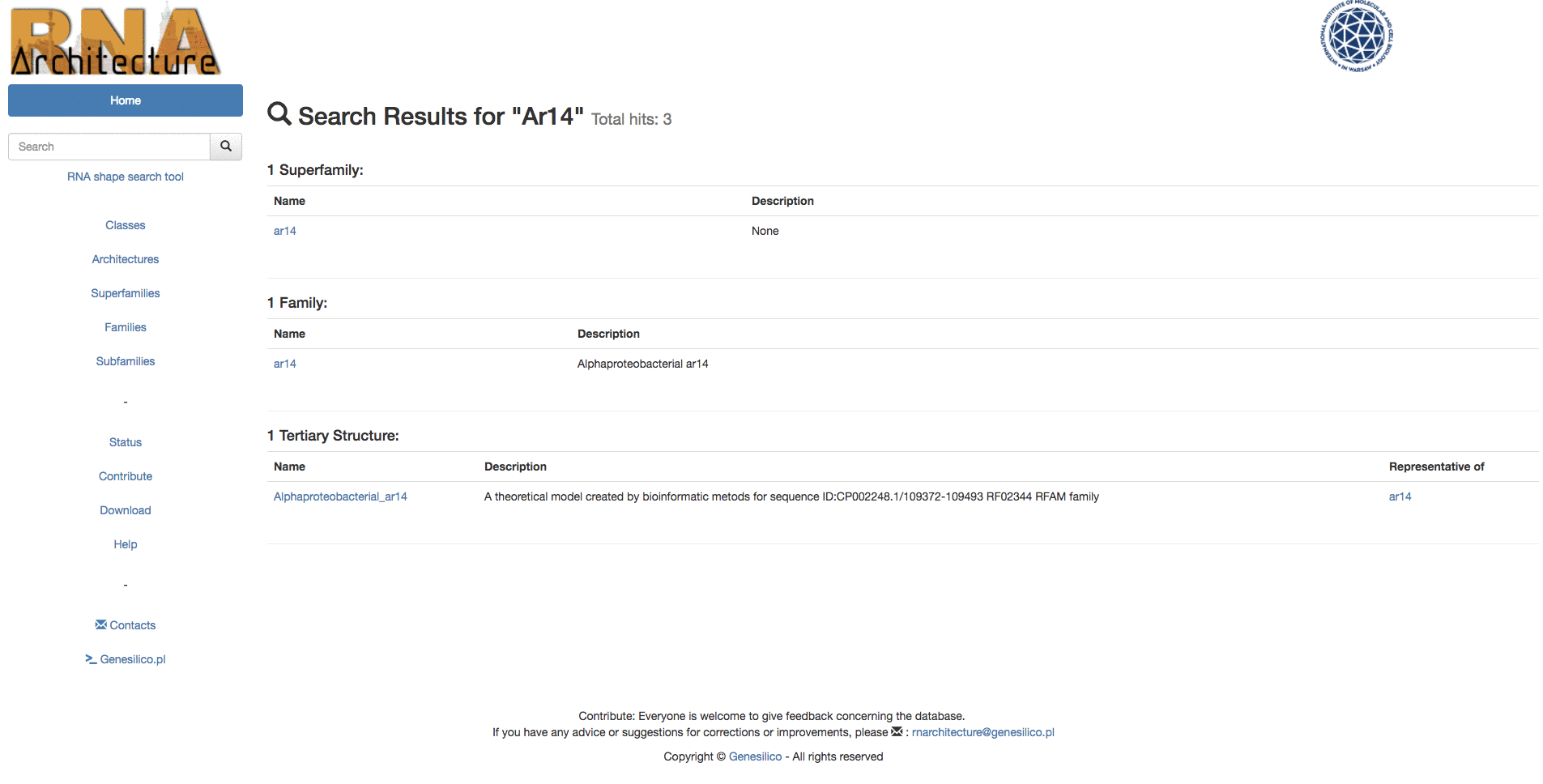 Example: Searching for "Intron" keyword from the homepage, then using the left menu search-bar to search "Ar14". The two search bars are equivalents.
Example: Searching for "Intron" keyword from the homepage, then using the left menu search-bar to search "Ar14". The two search bars are equivalents.
To improve the usability of the RNArchitecture database we have extended the search utility to include the RNA shapes. The user can now reveal all RNA families that possess a particular architectural motif (e.g. a simple pseudoknot) by querying the database with “([)]”. Currently such searches are either not possible or very difficult to make with other databases of RNA families or RNA structures.
To access the RNA Shape search tool, click on the link below the search bar in the left menu bar (Fig.4).
 Fig. 4 - Position of the link to access to the RNA Shape search tool
Fig. 4 - Position of the link to access to the RNA Shape search tool
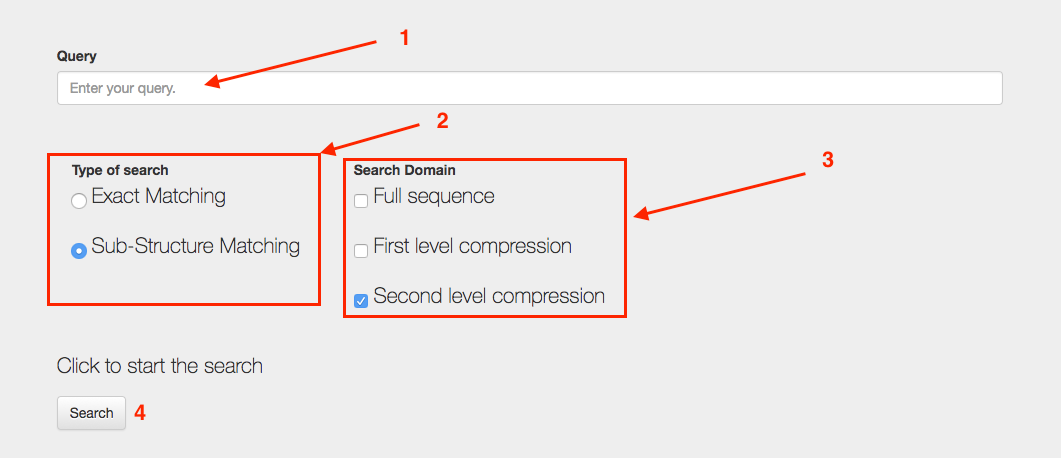 Fig. 5 - RNA Shape search tool search process and query interface
Fig. 5 - RNA Shape search tool search process and query interface
Fig. 5 - point 1. THE QUERY
To perform a search insert your query in the query text box. It's possible to use the following symbols to perform the query : . , & , ( ) , [ ] , { } , < >. If other characters are used the search will return no results.
Fig. 5 - point 2. TYPE OF SEARCH
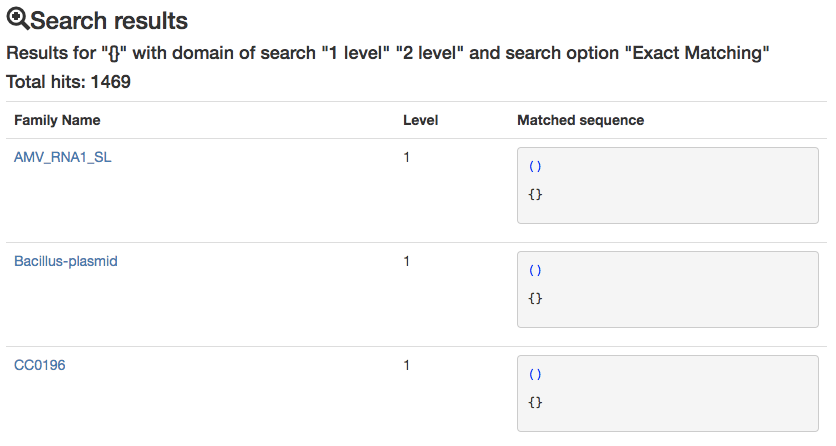
Select the type of search that you would like to perform.The first option, Exact match returns only the families which secondary structure topology match exactly the query.
Example: the query () will match the following topologies {} [] () and <> but will exclude all the families where the submotif is present but not unique.
The second option Sub-structure marching offers instead the possibility of exploring the sub-structures contained inside a longer sequence The queried topology is matched even more times if found in a different position of the same secondary sequence structure. Example: again, for the query (), this time the results would match all the Sub-Structures in few families sequences:
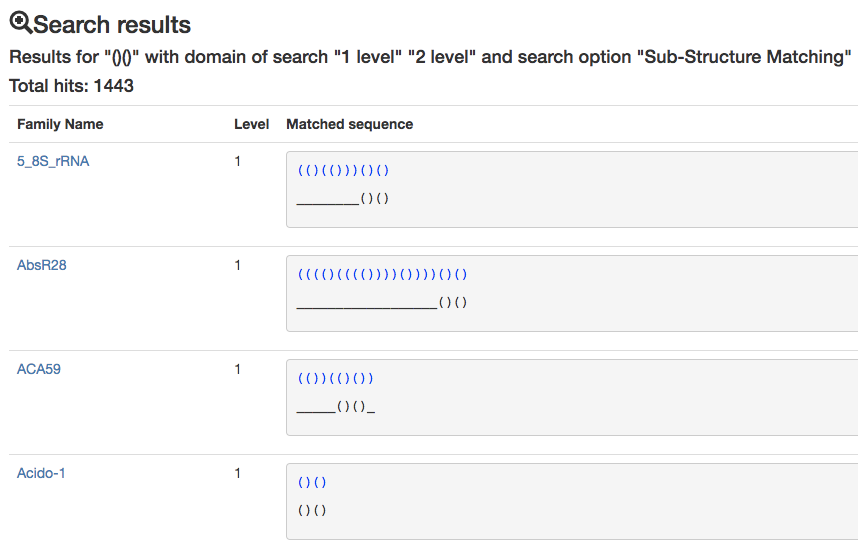
Fig. 5 - point 3. DOMAIN OF SEARCH
The domain of search option defines which entries of the databases will be explored by the search engine. There are three possibilities. Full sequence, first level compression or second level compression search.
Fig. 5 - point 4. SEARCH
Once all your search is set up you can press the button "Search" to query the database and receive the results.
In the following animation you have an example of how to perform two RNA shape search queries on the database. The first query is an exact match, while the second is a sub-structure search.