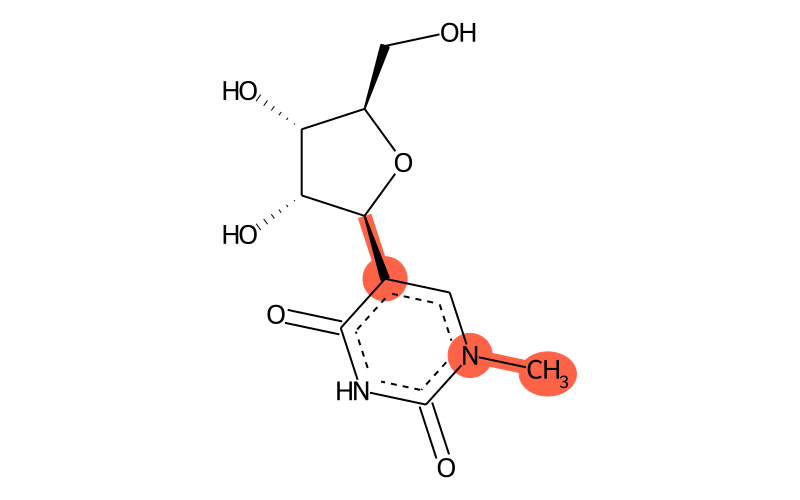
Summary
| Full name | 1-methylpseudouridine |
| IUPAC name | 5-[(2S,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-1-methylpyrimidine-2,4-dione |
| Short name | m1Y |
| MODOMICS code new | 2000000019U |
| MODOMICS code | 19U |
| Synonyms |
09RAD4M6WF
13860-38-3 1-methyl-pseudouridine 1-Methylpseudouridine 1-N-Methyl-pseudouridine (1S)-1,4-anhydro-1-(1-methyl-2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)-D-ribitol 2,4(1H,3H)-Pyrimidinedione, 1-methyl-5-beta-D-ribofuranosyl- 2,4(1H,3H)-Pyrimidinedione,1-methyl-5-b-D-ribofuranosyl- 5-[(2S,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-1-methylpyrimidine-2,4-dione 5-((2S,3R,4S,5R)-3,4-Dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl)-1-methylpyrimidine-2,4(1H,3H)-dione AC1L40YV AKOS030241474 C10-H14-N2-O6 CHEBI:19068 CID99543 CS-0047598 CTK4C1317 DTXSID50160724 HY-112582 m(1)f MFCD10687017 N1-methyl-pseudouridine N1-Methylpseudouridine NSC 240023 Q27109108 SCHEMBL63879 U-50228 UNII-09RAD4M6WF ZINC6072377 |
| Nature of the modified residue | Natural |
| RNAMods code | ] |
| Residue unique ID | 88 |
| Found in RNA | Yes |
| Related nucleotides | 345 |
| Enzymes |
Nep1 (Saccharomyces cerevisiae) Nep1 (Methanocaldococcus jannaschii) TrmY (Haloferax volcanii) TrmY (Methanocaldococcus jannaschii) |
| Found in phylogeny | Archaea, Eukaryota |
| Found naturally in RNA types | rRNA, tRNA |
Chemical information
| Sum formula | C10H14N2O6 |
| Type of moiety | nucleoside |
| Degeneracy | not applicable |
| SMILES | C[n]1c(=O)[nH]c(=O)c([C@H]2[C@H](O)[C@H](O)[C@@H](CO)O2)c1 |
| logP | -2.7724 |
| TPSA | 124.78 |
| Number of atoms | 18 |
| Number of Hydrogen Bond Acceptors 1 (HBA1) | 6 |
| Number of Hydrogen Bond Acceptors 2 (HBA2) | 7 |
| Number of Hydrogen Bond Donors (HBD) | 4 |
| PDB no exac match , link to the most similar ligand | 5MD |
| HMDB (Human Metabolome Database) no exact match, link to the most similar ligand | None |
| InChI | InChI=1S/C10H14N2O6/c1-12-2-4(9(16)11-10(12)17)8-7(15)6(14)5(3-13)18-8/h2,5-8,13-15H,3H2,1H3,(H,11,16,17)/t5-,6-,7-,8+/m1/s1 |
| InChIKey | UVBYMVOUBXYSFV-XUTVFYLZSA-N |
| Search the molecule in external databases | ChEMBL ChemAgora ChEBI PubChem Compound Database Ligand Expo ChemSpider WIPO |
| PubChem CID | |
| PubChem SIDs |
10230648
44429739 57336910 75277141 104424084 135057577 135668181 162789635 163704614 226445208 257913638 273527963 312237482 315721216 319133262 337257376 341192126 349655324 355073765 363671132 375974003 381188910 383857692 384240268 385541565 385663805 386500321 386978219 387065207 388915103 404608685 405258086 405268277 419580329 434720109 439435858 442077450 |
* Chemical properties calculated with Open Babel - O'Boyle et al. Open Babel: An open chemical toolbox. J Cheminform 3, 33 (2011) (link)
Download Structures
| 2D | .png .mol .mol2 .sdf .pdb .smi |
| 3D | .mol .mol2 .sdf .pdb |
Tautomers
| Tautomers SMILES |
Cn1c(=O)[nH]c(=O)c(C2C(O)C(O)C(CO)O2)c1 tautomer #0
Cn1c(O)nc(=O)c(C2C(O)C(O)C(CO)O2)c1 tautomer #1 Cn1c(=O)nc(O)c(C2C(O)C(O)C(CO)O2)c1 tautomer #2 |
| Tautomer image | Show Image |
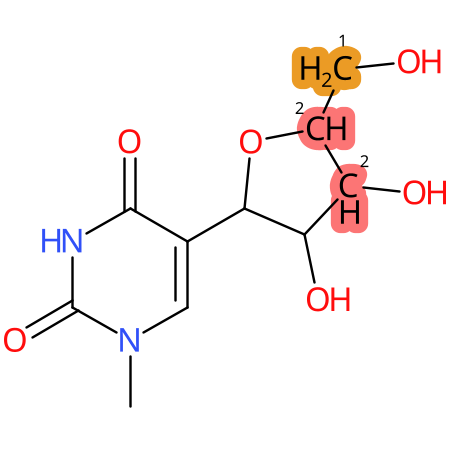
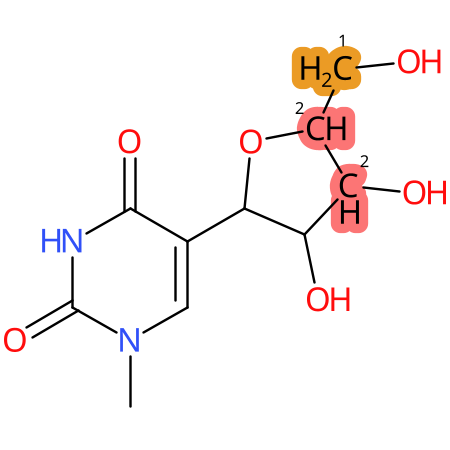
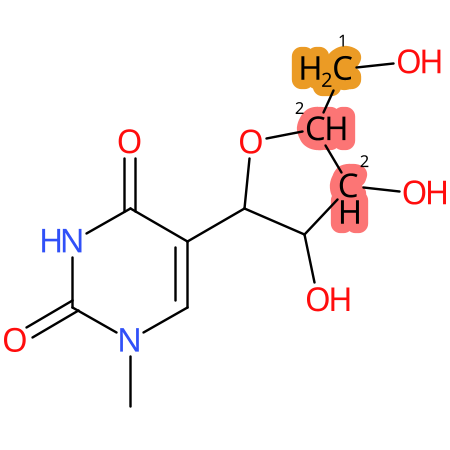
Predicted CYP Metabolic Sites
| CYP3A4 | CYP2D6 | CYP2C9 |
|---|---|---|
|
|
|
* CYP Metabolic sites predicted with SMARTCyp. SMARTCyp is a method for prediction of which sites in a molecule that are most liable to metabolism by Cytochrome P450. It has been shown to be applicable to metabolism by the isoforms 1A2, 2A6, 2B6, 2C8, 2C19, 2E1, and 3A4 (CYP3A4), and specific models for the isoform 2C9 (CYP2C9) and isoform 2D6 (CYP2D6). CYP3A4, CYP2D6, and CYP2C9 are the three of the most important enzymes in drug metabolism since they are involved in the metabolism of more than half of the drugs used today. The three top-ranked atoms are highlighted. See: SmartCYP and SmartCYP - background; Patrik Rydberg, David E. Gloriam, Lars Olsen, The SMARTCyp cytochrome P450 metabolism prediction server, Bioinformatics, Volume 26, Issue 23, 1 December 2010, Pages 2988–2989 (link)
LC-MS Information
| Monoisotopic mass | 258.0852 |
| Average mass | 258.228 |
| [M+H]+ | 259.093 |
| Product ions | 169/179/227/209 |
| Normalized LC elution time * | not available |
| LC elution order/characteristics | not available |
* normalized to guanosine (G), measured with a RP C-18 column with acetonitrile/ammonium acetate as mobile phase.
Chemical groups contained
| Type | Subtype |
|---|---|
| methyl group | methyl at aromatic N |
| ring modification | pseudouridine |
Reactions producing 1-methylpseudouridine
| Name |
|---|
| Y:m1Y |
Reactions starting from 1-methylpseudouridine
| Name |
|---|
| m1Y:m1acp3Y |
Last modification of this entry: Sept. 22, 2023