Summary
| Full name | 7-aminomethyl-7-carbaguanine |
| IUPAC name | 2-amino-5-(aminomethyl)-3,7-dihydropyrrolo[2,3-d]pyrimidin-4-one |
| Short name | preQ1base |
| MODOMICS code new | 2000101000G |
| MODOMICS code | 101000G |
| Synonyms |
2-Amino-5-({[(1s,4s,5r)-4,5-Dihydroxycyclopent-2-En-1-Yl]amino}methyl)-3,7-Dihydro-4h-Pyrrolo[2,3-D]pyrimidin-4-One
2-amino-5-(aminomethyl)-1,7-dihydro-4h-pyrrolo[2,3-d]pyrimidin-4-one 2-amino-5-(aminomethyl)-1,7-dihydropyrrolo[2,3-d]pyrimidin-4-one 2-amino-5-(aminomethyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one 2-amino-5-(aminomethyl)-3H,4H,7H-pyrrolo[2,3-d]pyrimidin-4-one 2-Amino-5-(aminomethyl)-3H-pyrrolo[2,3-d]pyrimidin-4(7H)-one 2-Amino-5-(aminomethyl)pyrrolo[2,3-d]pyrimidin-4(3H)-one 69251-45-2 7-Aminomethyl-7-carbaguanine 7-aminomethyl-7-deazaguanine 7-DEAZA-7-AMINOMETHYL-GUANINE AC1L18MX C16675 CHEBI:45126 CHEMBL1235432 CID171 CTK7E3986 DB03304 HNG NS00069406 preQ1 PRF Q27120591 QEI queuine SCHEMBL166747 SCHEMBL20206657 ZINC3869370 |
| Nature of the modified residue | Natural |
| RNAMods code | ∇ |
| Residue unique ID | 61 |
| Found in RNA | Yes |
| Enzymes |
QueF (Escherichia coli) QueF (Bacillus subtilis) |
| Found in phylogeny | Eubacteria |
Chemical information
| Sum formula | C7H9N5O |
| Type of moiety | base |
| Degeneracy | not applicable |
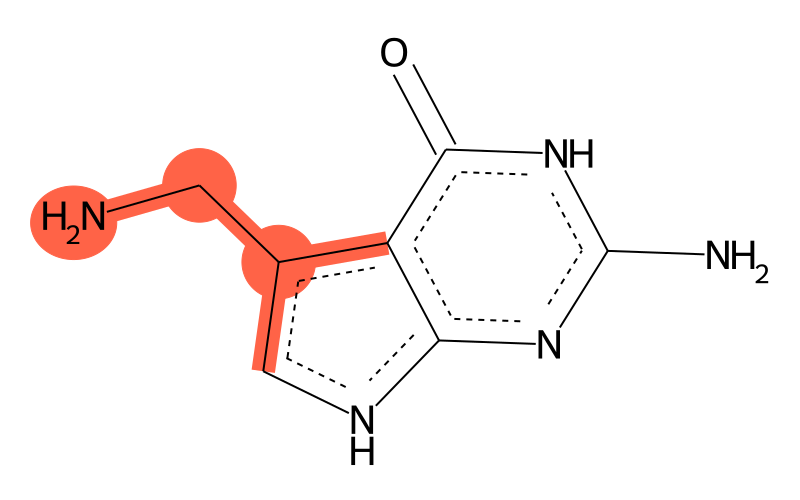
| SMILES | Nc1[nH]c(=O)c2c([nH]cc2CN)n1 |
| logP | 0.5736 |
| TPSA | 113.58 |
| Number of atoms | 13 |
| Number of Hydrogen Bond Acceptors 1 (HBA1) | 4 |
| Number of Hydrogen Bond Acceptors 2 (HBA2) | 4 |
| Number of Hydrogen Bond Donors (HBD) | 4 |
| PDB no exac match , link to the most similar ligand | HNG |
| PDB exact match , link to the most similar ligand | PRF |
| HMDB (Human Metabolome Database) exact match | None |
| HMDB (Human Metabolome Database) no exact match, link to the most similar ligand | None |
| InChI | InChI=1S/C7H9N5O/c8-1-3-2-10-5-4(3)6(13)12-7(9)11-5/h2H,1,8H2,(H4,9,10,11,12,13) |
| InChIKey | MEYMBLGOKYDGLZ-UHFFFAOYSA-N |
| Search the molecule in external databases | ChEMBL ChemAgora ChEBI PubChem Compound Database Ligand Expo ChemSpider WIPO |
| PubChem CID | |
| PubChem SIDs |
829087
829100 829910 838254 4399942 7890044 24438548 29195969 29196259 46506298 51090998 56343614 56435741 57280190 57304842 57320008 75058265 85240117 99289336 104251581 104294054 121360469 124361858 128585993 144072512 144080810 152255351 160650696 162010276 162108542 163018900 172112764 226528764 249765068 252082309 257081220 274335793 293094581 312227373 316907842 348281437 355196640 369381740 374823142 376920328 381339551 381999846 383859890 384992625 385664864 386501384 387123075 393805940 405580822 419572099 443408118 |
* Chemical properties calculated with Open Babel - O'Boyle et al. Open Babel: An open chemical toolbox. J Cheminform 3, 33 (2011) (link)
Download Structures
| 2D | .png .mol .mol2 .sdf .pdb .smi |
| 3D | .mol .mol2 .sdf .pdb |
Tautomers
| Tautomers SMILES |
N=C1NC(O)c2c([nH]cc2C=N)N1 tautomer #0
N=c1[nH]c(=O)c2c([nH]cc2CN)[nH]1 tautomer #1 NC=1NC(O)c2c([nH]cc2C=N)N1 tautomer #2 Nc1nc(O)c2c([nH]cc2CN)n1 tautomer #3 NC1=NC(O)c2c([nH]cc2C=N)N1 tautomer #4 Nc1nc(O)c2-c(ncc2CN)[nH]1 tautomer #5 Nc1[nH]c(O)c2-c(ncc2CN)n1 tautomer #6 Nc1nc(=O)c2c([nH]cc2CN)[nH]1 tautomer #7 Nc1[nH]c(=O)c2c([nH]cc2CN)n1 tautomer #8 Nc1nc(O)c2c(NCC2C=N)n1 tautomer #9 Nc1nc(O)c2c(N=CC2CN)n1 tautomer #10 Nc1nc(O)c2c(NCC2=CN)n1 tautomer #11 N=c1[nH]c(O)c2c([nH]cc2CN)n1 tautomer #12 N=C1NC(=O)C2C(N=CC2C=N)N1 tautomer #13 N=C1NC(=O)C=2C(N=CC2CN)N1 tautomer #14 N=C1NC(O)C=2C(N=CC2C=N)N1 tautomer #15 N=C1NC(=O)C=2C(NCC2C=N)N1 tautomer #16 N=C1NC(O)C=2C(N=CC2CN)=N1 tautomer #17 N=C1NC(=O)C2C(NCC2C=N)=N1 tautomer #18 N=C1NC(O)C=2C(NCC2C=N)=N1 tautomer #19 N=C1NC(=O)C=2C(NCC2CN)=N1 tautomer #20 N=C1NC(=O)C2C(N=CC2CN)=N1 tautomer #21 NC1=NC(O)C2C(N=CC2C=N)=N1 tautomer #22 NC=1NC(=O)C=2C(NCC2C=N)N1 tautomer #23 NC=1NC(O)C=2C(=NCC2C=N)N1 tautomer #24 Nc1[nH]c(=O)c=2c(=NCC2CN)n1 tautomer #25 NC=1NC(=O)C=2C(N=CC2CN)N1 tautomer #26 NC=1NC(=O)C2C(N=CC2C=N)N1 tautomer #27 NC=1NC(O)C=2C(N=CC2C=N)N1 tautomer #28 N=C1NC(=O)C2C(NC=C2C=N)N1 tautomer #29 N=C1NC(=O)C2C(N=CC2=CN)N1 tautomer #30 N=C1NC(=O)C2C(NC=C2CN)=N1 tautomer #31 N=C1NC(=O)C2C(NCC2=CN)=N1 tautomer #32 N=C1NC(O)C2=C(N=CC2C=N)N1 tautomer #33 N=c1[nH]c(=O)c2c(NCC2C=N)[nH]1 tautomer #34 NC1=NC(O)C2C(N=CC2=CN)=N1 tautomer #35 NC1=NC(O)C2C(NC=C2C=N)=N1 tautomer #36 Nc1[nH]c(=O)c2c(NCC2C=N)n1 tautomer #37 NC=1NC(O)C2=C(N=CC2C=N)N1 tautomer #38 NC=1NC(=O)C2C(N=CC2=CN)N1 tautomer #39 NC=1NC(=O)C2C(NC=C2C=N)N1 tautomer #40 Nc1nc(O)c=2c(=NCC2CN)n1 tautomer #41 N=C1NC(O)C2=C(N=CC2=CN)N1 tautomer #42 N=c1[nH]c(=O)c2c(NCC2=CN)[nH]1 tautomer #43 N=c1[nH]c(=O)c2c(N=CC2CN)[nH]1 tautomer #44 Nc1[nH]c(=O)c2c(NCC2=CN)n1 tautomer #45 NC=1NC(O)C2=C(N=CC2=CN)N1 tautomer #46 Nc1nc(O)c2c(NCC2C=N)n1 tautomer #47 Nc1nc(O)c2c([nH]cc2CN)n1 tautomer #48 NC1=NC(=O)C2C(N=CC2C=N)N1 tautomer #49 NC1=NC(=O)C=2C(N=CC2CN)N1 tautomer #50 NC1=NC(O)C=2C(N=CC2C=N)N1 tautomer #51 NC1=NC(=O)C=2C(NCC2C=N)N1 tautomer #52 NC1=NC(O)C=2C(N=CC2CN)=N1 tautomer #53 NC1=NC(=O)C2C(NCC2C=N)=N1 tautomer #54 NC1=NC(O)C=2C(NCC2C=N)=N1 tautomer #55 NC1=NC(=O)C=2C(NCC2CN)=N1 tautomer #56 N=C1NC(=O)C2C(=NCC2C=N)N1 tautomer #57 N=C1NC(O)C=2C(=NCC2C=N)N1 tautomer #58 N=c1[nH]c(=O)c=2c(=NCC2CN)[nH]1 tautomer #59 NC1=NC(=O)C2C(N=CC2CN)=N1 tautomer #60 N=C1NC(O)C2C(N=CC2C=N)=N1 tautomer #61 Nc1nc(O)c2c(NCC2=CN)n1 tautomer #62 NC1=NC(=O)C2C(NC=C2C=N)N1 tautomer #63 NC1=NC(=O)C2C(N=CC2=CN)N1 tautomer #64 NC1=NC(=O)C2C(NC=C2CN)=N1 tautomer #65 NC1=NC(=O)C2C(NCC2=CN)=N1 tautomer #66 NC1=NC(O)C2=C(N=CC2C=N)N1 tautomer #67 N=C1NC(=O)C2C(=NC=C2CN)N1 tautomer #68 N=C1NC(=O)C2C(=NCC2=CN)N1 tautomer #69 N=C1NC(O)C2C(N=CC2=CN)=N1 tautomer #70 N=C1NC(O)C2C(NC=C2C=N)=N1 tautomer #71 Nc1nc(=O)c2c(NCC2C=N)[nH]1 tautomer #72 NC1=NC(O)C2=C(N=CC2=CN)N1 tautomer #73 Nc1nc(=O)c2c(N=CC2CN)[nH]1 tautomer #74 Nc1nc(=O)c2c(NCC2=CN)[nH]1 tautomer #75 NC1=NC(=O)C2C(=NCC2C=N)N1 tautomer #76 NC1=NC(O)C=2C(=NCC2C=N)N1 tautomer #77 Nc1nc(=O)c=2c(=NCC2CN)[nH]1 tautomer #78 NC=1NC(=O)C2C(=NC=C2CN)N1 tautomer #79 NC=1NC(=O)C2C(=NCC2C=N)N1 tautomer #80 N=c1nc(O)c2c([nH]cc2CN)[nH]1 tautomer #81 NC1=NC(=O)C2C(=NC=C2CN)N1 tautomer #82 NC1=NC(=O)C2C(=NCC2=CN)N1 tautomer #83 Nc1[nH]c(=O)c2c(N=CC2CN)n1 tautomer #84 NC=1NC(=O)C2C(=NCC2=CN)N1 tautomer #85 NC=1NC(O)=C2C(=NCC2C=N)N1 tautomer #86 N=C1NC(O)=C2C(N=CC2C=N)N1 tautomer #87 N=c1[nH]c(O)c2c(NCC2C=N)n1 tautomer #88 N=c1[nH]c(O)c2c(N=CC2CN)n1 tautomer #89 NC=1NC(O)=C2C(=NCC2=CN)N1 tautomer #90 NC=1NC(O)=C2C(N=CC2C=N)N1 tautomer #91 Nc1nc(O)c2c(N=CC2CN)n1 tautomer #92 N=C1NC(O)=C2C(N=CC2=CN)N1 tautomer #93 NC=1NC(O)=C2C(N=CC2=CN)N1 tautomer #94 NC=1NC(O)=C2C(NC=C2C=N)N1 tautomer #95 N=C1NC(O)=C2C(NC=C2C=N)N1 tautomer #96 N=c1[nH]c(O)c2c(NCC2=CN)n1 tautomer #97 NC1=NC(O)=C2C(N=CC2C=N)N1 tautomer #98 NC1=NC(O)=C2C(NC=C2C=N)N1 tautomer #99 NC1=NC(O)=C2C(N=CC2=CN)N1 tautomer #100 N=C1NC(O)=C2C(=NCC2C=N)N1 tautomer #101 N=c1[nH]c(O)c2-c(ncc2CN)[nH]1 tautomer #102 N=C1NC(O)=C2C(=NCC2=CN)N1 tautomer #103 NC1=NC(O)=C2C(=NCC2C=N)N1 tautomer #104 NC1=NC(O)=C2C(=NCC2=CN)N1 tautomer #105 N=C1N=C(O)C2C(N=CC2C=N)N1 tautomer #106 N=C1N=C(O)C=2C(N=CC2CN)N1 tautomer #107 N=C1N=C(O)C=2C(NCC2C=N)N1 tautomer #108 N=C1N=C(O)C2C(NCC2C=N)=N1 tautomer #109 N=C1N=C(O)C=2C(NCC2CN)=N1 tautomer #110 N=C1N=C(O)C2C(N=CC2CN)=N1 tautomer #111 NC=1N=C(O)C=2C(NCC2C=N)N1 tautomer #112 NC=1N=C(O)C=2C(N=CC2CN)N1 tautomer #113 NC=1N=C(O)C2C(N=CC2C=N)N1 tautomer #114 N=C1N=C(O)C2C(NC=C2C=N)N1 tautomer #115 N=C1N=C(O)C2C(N=CC2=CN)N1 tautomer #116 N=C1N=C(O)C2C(NC=C2CN)=N1 tautomer #117 N=C1N=C(O)C2C(NCC2=CN)=N1 tautomer #118 NC=1N=C(O)C2C(N=CC2=CN)N1 tautomer #119 NC=1N=C(O)C2C(NC=C2C=N)N1 tautomer #120 N=c1nc(O)c2c(NCC2C=N)[nH]1 tautomer #121 N=c1nc(O)c2c(N=CC2CN)[nH]1 tautomer #122 N=c1nc(O)c2c(NCC2=CN)[nH]1 tautomer #123 N=C1N=C(O)C2C(=NCC2C=N)N1 tautomer #124 N=c1nc(O)c=2c(=NCC2CN)[nH]1 tautomer #125 NC=1N=C(O)C2C(=NCC2C=N)N1 tautomer #126 N=C1N=C(O)C2C(=NC=C2CN)N1 tautomer #127 N=C1N=C(O)C2C(=NCC2=CN)N1 tautomer #128 NC=1N=C(O)C2C(=NCC2=CN)N1 tautomer #129 NC=1N=C(O)C2C(=NC=C2CN)N1 tautomer #130 |
| Tautomer image | Show Image |
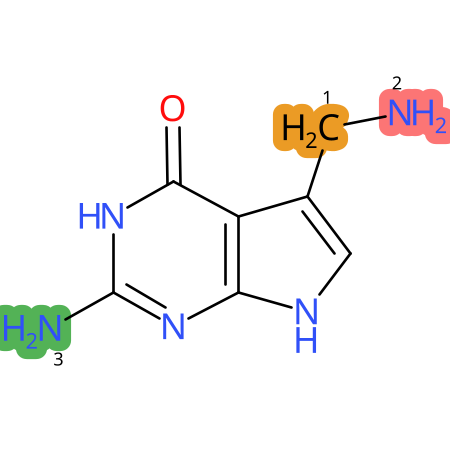
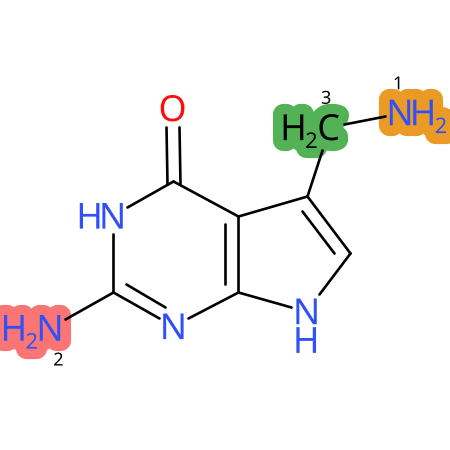
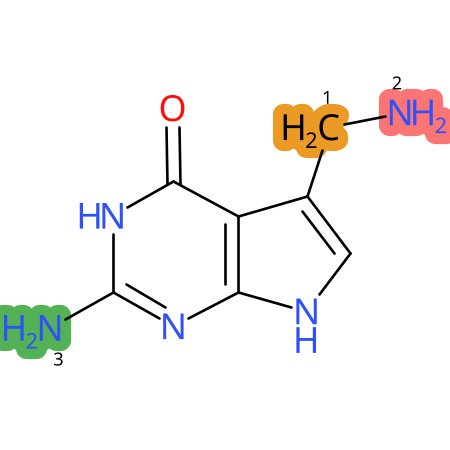
Predicted CYP Metabolic Sites
| CYP3A4 | CYP2D6 | CYP2C9 |
|---|---|---|
|
|
|
* CYP Metabolic sites predicted with SMARTCyp. SMARTCyp is a method for prediction of which sites in a molecule that are most liable to metabolism by Cytochrome P450. It has been shown to be applicable to metabolism by the isoforms 1A2, 2A6, 2B6, 2C8, 2C19, 2E1, and 3A4 (CYP3A4), and specific models for the isoform 2C9 (CYP2C9) and isoform 2D6 (CYP2D6). CYP3A4, CYP2D6, and CYP2C9 are the three of the most important enzymes in drug metabolism since they are involved in the metabolism of more than half of the drugs used today. The three top-ranked atoms are highlighted. See: SmartCYP and SmartCYP - background; Patrik Rydberg, David E. Gloriam, Lars Olsen, The SMARTCyp cytochrome P450 metabolism prediction server, Bioinformatics, Volume 26, Issue 23, 1 December 2010, Pages 2988–2989 (link)
LC-MS Information
| Monoisotopic mass | 179.0807 |
| Average mass | 179.179 |
| [M+H]+ | not available |
| Product ions | not available |
| Normalized LC elution time * | not available |
| LC elution order/characteristics | not available |
* normalized to guanosine (G), measured with a RP C-18 column with acetonitrile/ammonium acetate as mobile phase.
Chemical groups contained
| Type | Subtype |
|---|---|
| methyl group | methyl at aromatic C |
| other | primary amine |
Reactions producing 7-aminomethyl-7-carbaguanine
| Name |
|---|
| preQ0base:preQ1base |
Reactions starting from 7-aminomethyl-7-carbaguanine
| Name |
|---|
| preQ1base:preQ1 |
Last modification of this entry: Oct. 9, 2023