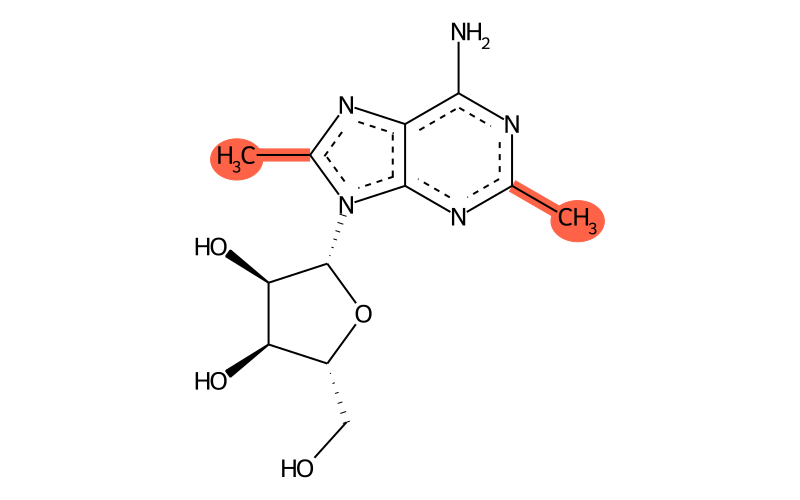
Summary
| Full name | 2,8-dimethyladenosine |
| IUPAC name | (2R,3R,4S,5R)-2-(6-amino-2,8-dimethylpurin-9-yl)-5-(hydroxymethyl)oxolane-3,4-diol |
| Short name | m2,8A |
| MODOMICS code new | 2000000028A |
| MODOMICS code | 28A |
| Synonyms |
2,8-dimethyladenosine
(2R,3R,4S,5R)-2-(6-Amino-2,8-dimethyl-9H-purin-9-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol (2R,3R,4S,5R)-2-(6-amino-2,8-dimethylpurin-9-yl)-5-(hydroxymethyl)oxolane-3,4-diol 63954-66-5 SCHEMBL18740403 |
| Nature of the modified residue | Natural |
| RNAMods code | ± |
| Residue unique ID | 136 |
| Found in RNA | Yes |
| Related nucleotides | 371 |
| Enzymes |
Cfr (Staphylococcus sciuri) |
| Found in phylogeny | Eubacteria |
| Found naturally in RNA types | rRNA |
Chemical information
| Sum formula | C12H17N5O4 |
| Type of moiety | nucleoside |
| Degeneracy | not applicable |
| SMILES | Cc1nc2c(N)nc(C)nc2[n]1[C@@H]1O[C@H](CO)[C@@H](O)[C@H]1O |
| logP | -0.782 |
| TPSA | 139.54 |
| Number of atoms | 21 |
| Number of Hydrogen Bond Acceptors 1 (HBA1) | 8 |
| Number of Hydrogen Bond Acceptors 2 (HBA2) | 9 |
| Number of Hydrogen Bond Donors (HBD) | 4 |
| PDB no exac match , link to the most similar ligand | N8M |
| HMDB (Human Metabolome Database) no exact match, link to the most similar ligand | None |
| InChI | InChI=1S/C12H17N5O4/c1-4-14-10(13)7-11(15-4)17(5(2)16-7)12-9(20)8(19)6(3-18)21-12/h6,8-9,12,18-20H,3H2,1-2H3,(H2,13,14,15)/t6-,8-,9-,12-/m1/s1 |
| InChIKey | FGMBEEFIKCGALL-WOUKDFQISA-N |
| Search the molecule in external databases | ChEMBL ChemAgora ChEBI PubChem Compound Database Ligand Expo ChemSpider WIPO |
| PubChem CID | |
| PubChem SIDs |
* Chemical properties calculated with Open Babel - O'Boyle et al. Open Babel: An open chemical toolbox. J Cheminform 3, 33 (2011) (link)
Download Structures
| 2D | .png .mol .mol2 .sdf .pdb .smi |
| 3D | .mol .mol2 .sdf .pdb |
Tautomers
| Tautomers SMILES |
Cc1nc2c(N)nc(C)nc2n1C3OC(CO)C(O)C3O tautomer #0
Cc1nc2c(N)nc(C)nc2n1C3OC(CO)C(O)C3O tautomer #1 C=C1Nc2c(N)nc(C)nc2N1C3OC(CO)C(O)C3O tautomer #2 Cc1nc2c(=N)nc(C)[nH]c2n1C3OC(CO)C(O)C3O tautomer #3 Cc1nc2=C(N)NC(=C)N=c2n1C3OC(CO)C(O)C3O tautomer #4 Cc1nc2C(=N)NC(=C)Nc2n1C3OC(CO)C(O)C3O tautomer #5 C=C1Nc2c(N)nc(C)nc2N1C3OC(CO)C(O)C3O tautomer #6 Cc1nc2c(=N)[nH]c(C)nc2n1C3OC(CO)C(O)C3O tautomer #7 Cc1nc2C(N)=NC(=C)Nc2n1C3OC(CO)C(O)C3O tautomer #8 CC1=NC2C(=N)N=C(C)N=C2N1C3OC(CO)C(O)C3O tautomer #9 C=C1N=C2C(=N)N=C(C)NC2N1C3OC(CO)C(O)C3O tautomer #10 C=c1nc2C(N)N=C(C)N=c2n1C3OC(CO)C(O)C3O tautomer #11 CC1=NC2C(=N)NC(=C)N=C2N1C3OC(CO)C(O)C3O tautomer #12 C=C1NC2C(=N)N=C(C)N=C2N1C3OC(CO)C(O)C3O tautomer #13 C=C1NC2C(=N)NC(=C)N=C2N1C3OC(CO)C(O)C3O tautomer #14 C=c1nc2C(N)NC(=C)N=c2n1C3OC(CO)C(O)C3O tautomer #15 C=C1N=C2C(=N)NC(=C)NC2N1C3OC(CO)C(O)C3O tautomer #16 C=C1Nc2c(=N)nc(C)[nH]c2N1C3OC(CO)C(O)C3O tautomer #17 C=C1N=C2C(N)=NC(C)=NC2N1C3OC(CO)C(O)C3O tautomer #18 C=C1N=C2C(=N)NC(C)=NC2N1C3OC(CO)C(O)C3O tautomer #19 CC1=NC2C(N)=NC(=C)N=C2N1C3OC(CO)C(O)C3O tautomer #20 C=C1NC2=C(N)NC(=C)N=C2N1C3OC(CO)C(O)C3O tautomer #21 C=C1NC=2C(=N)NC(=C)NC2N1C3OC(CO)C(O)C3O tautomer #22 C=C1NC2C(N)=NC(=C)N=C2N1C3OC(CO)C(O)C3O tautomer #23 C=C1N=C2C(N)=NC(=C)NC2N1C3OC(CO)C(O)C3O tautomer #24 C=C1Nc2c(=N)[nH]c(C)nc2N1C3OC(CO)C(O)C3O tautomer #25 C=C1NC=2C(N)=NC(=C)NC2N1C3OC(CO)C(O)C3O tautomer #26 |
| Tautomer image | Show Image |
Predicted CYP Metabolic Sites
| CYP3A4 | CYP2D6 | CYP2C9 |
|---|---|---|
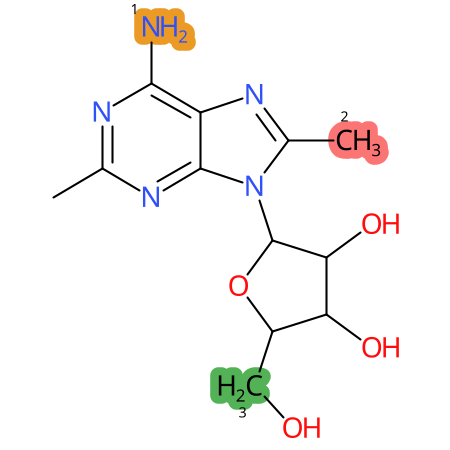
|
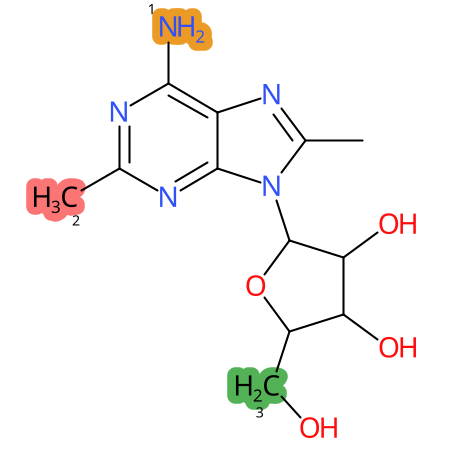
|
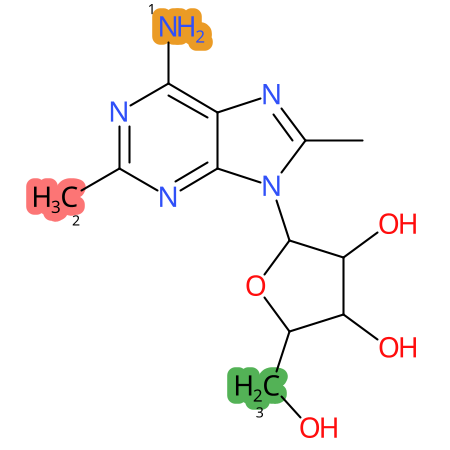
|
* CYP Metabolic sites predicted with SMARTCyp. SMARTCyp is a method for prediction of which sites in a molecule that are most liable to metabolism by Cytochrome P450. It has been shown to be applicable to metabolism by the isoforms 1A2, 2A6, 2B6, 2C8, 2C19, 2E1, and 3A4 (CYP3A4), and specific models for the isoform 2C9 (CYP2C9) and isoform 2D6 (CYP2D6). CYP3A4, CYP2D6, and CYP2C9 are the three of the most important enzymes in drug metabolism since they are involved in the metabolism of more than half of the drugs used today. The three top-ranked atoms are highlighted. See: SmartCYP and SmartCYP - background; Patrik Rydberg, David E. Gloriam, Lars Olsen, The SMARTCyp cytochrome P450 metabolism prediction server, Bioinformatics, Volume 26, Issue 23, 1 December 2010, Pages 2988–2989 (link)
LC-MS Information
| Monoisotopic mass | 295.1281 |
| Average mass | 295.294 |
| [M+H]+ | 296.1358 |
| Product ions | 164 |
| Normalized LC elution time * | not available |
| LC elution order/characteristics | not available |
* normalized to guanosine (G), measured with a RP C-18 column with acetonitrile/ammonium acetate as mobile phase.
Chemical groups contained
| Type | Subtype |
|---|---|
| methyl group | methyl group |
Reactions producing 2,8-dimethyladenosine
| Name |
|---|
| m2A:m2,8A |
| m8A:m2,8A |
Publications
| Title | Authors | Journal | Details | ||
|---|---|---|---|---|---|
| Identification of 8-methyladenosine as the modification catalyzed by the radical SAM methyltransferase Cfr that confers antibiotic resistance in bacteria. | Giessing AM, Jensen SS, Rasmussen A, Hansen LH, Gondela A, Long K, Vester B, Kirpekar F | RNA | [details] | 19144912 | - |
Last modification of this entry: Sept. 22, 2023