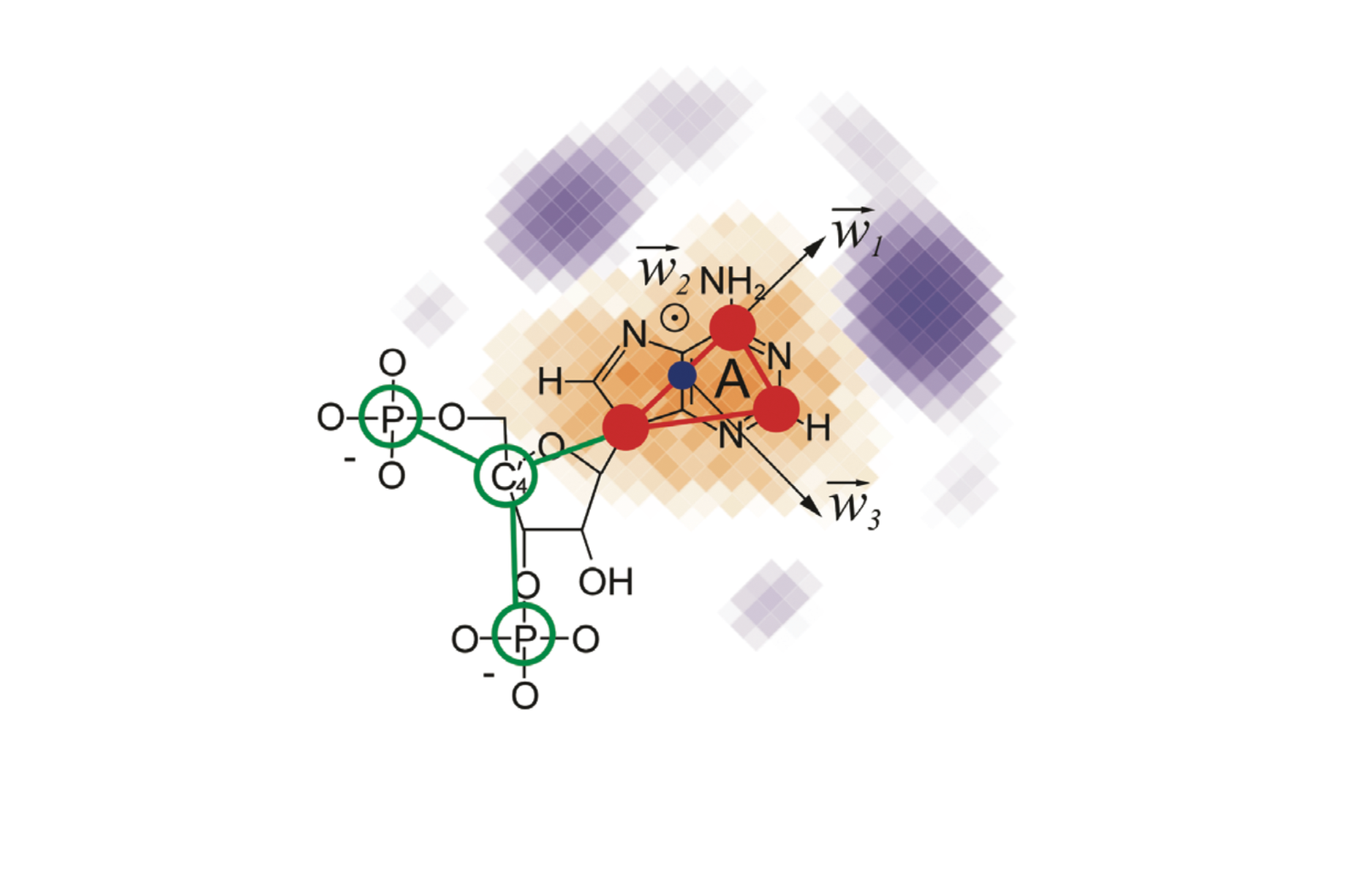
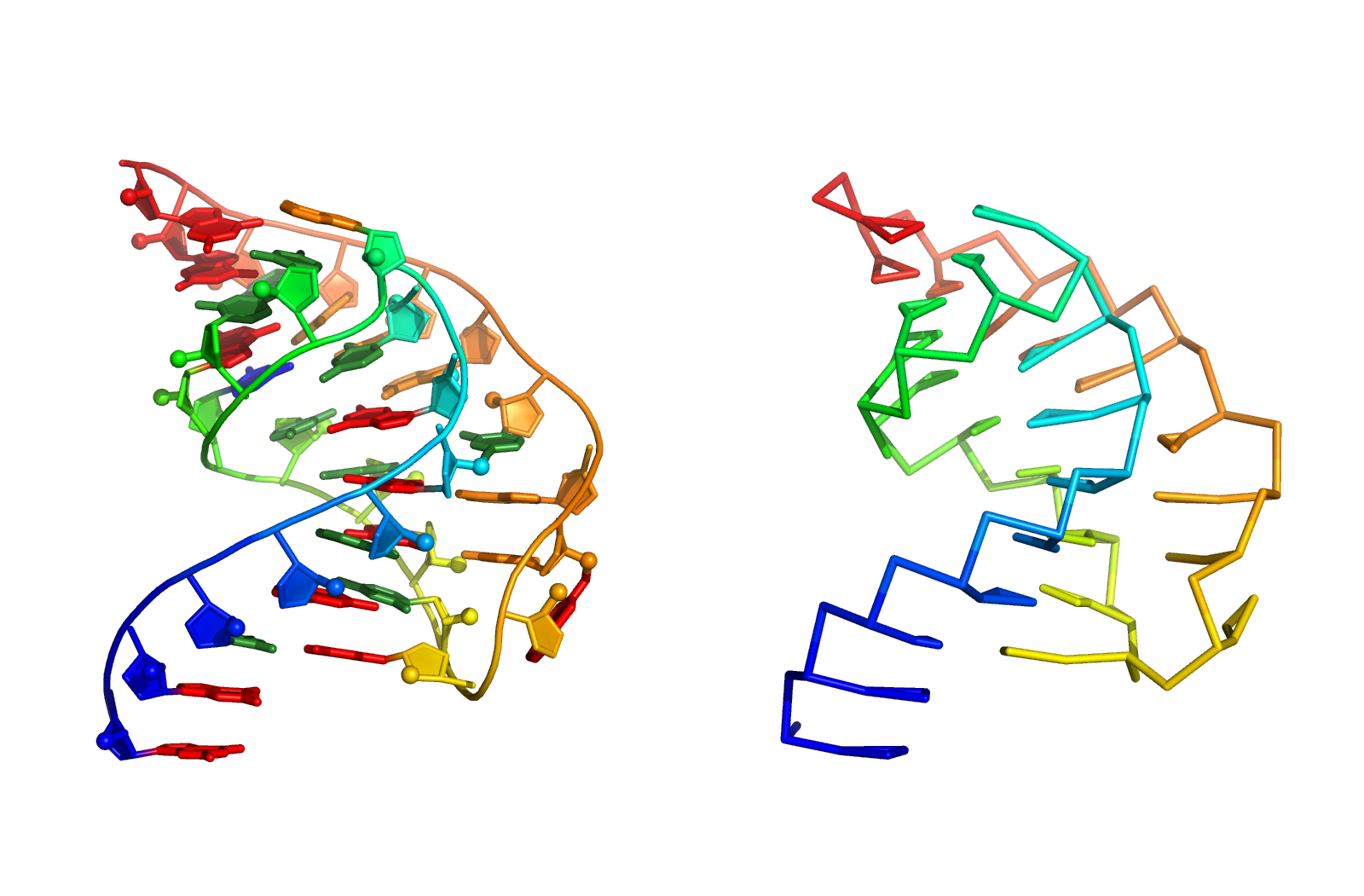
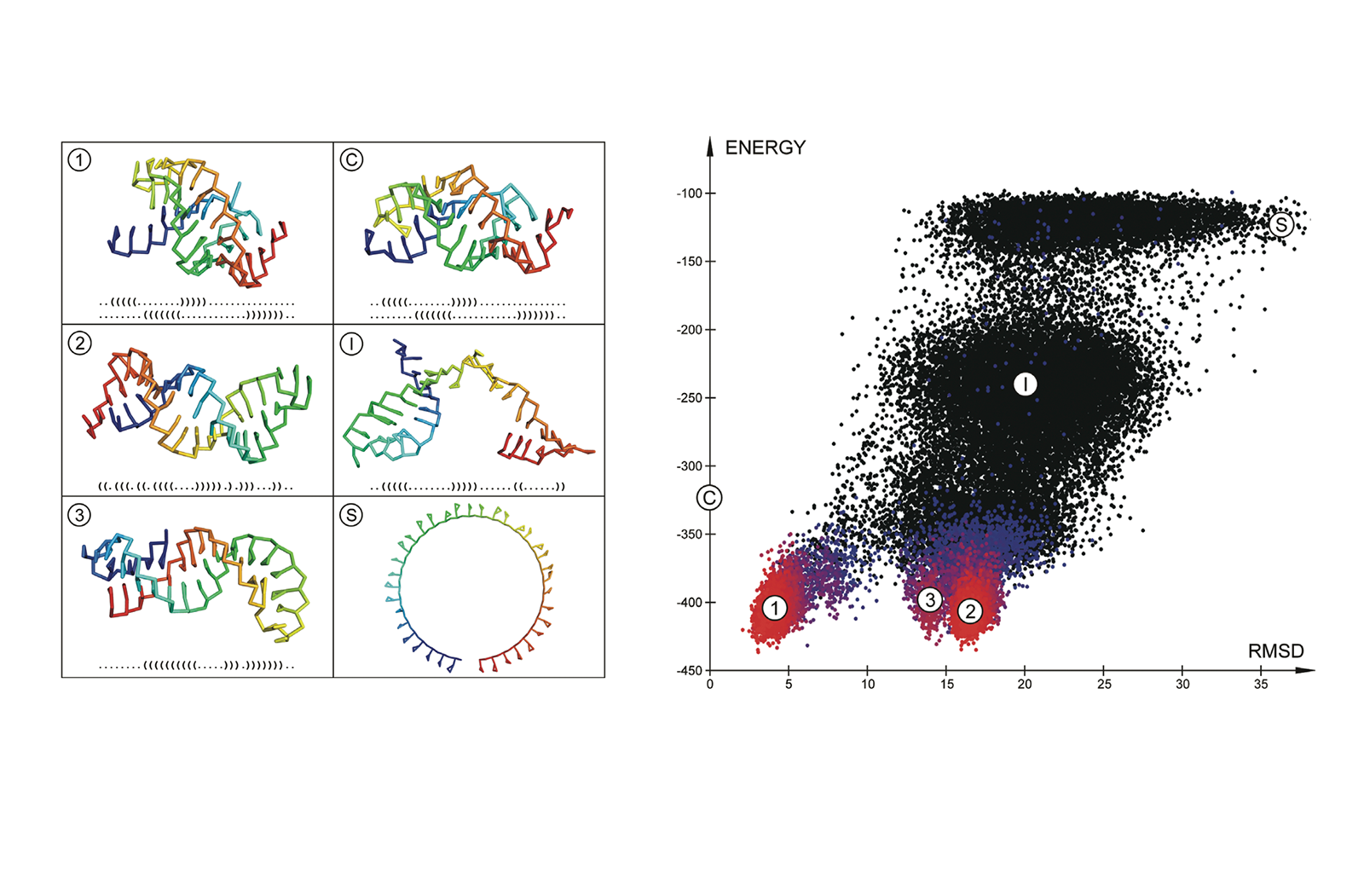
SimRNA is a method for RNA 3D structure modeling with optional restraints.
SimRNA uses (1) a coarse-grained representation, (2) relies on the Monte Carlo method for sampling the conformational space, and (3) employs a statistical potential to approximate the energy and identify conformations that correspond to biologically relevant structures.
What's new?
-
Incorporation of diverse restraints: Users can now implement a broader range of restraints, encompassing both canonical and non-canonical base pairs, and unpaired residues, as well as data from chemical probing experiments.
- Soft restraints on secondary structure [Dec. 2023]
- Hard restraints on base pairs [Dec. 2023]
- Soft restraints on base pairs [Dec. 2023]
- restraints on Chemical probing data [Dec. 2023]
- exclude Watson-Crick edge for base pairing [Dec. 2023]
- Advanced control over simulation parameters: A new way of defining restraints that can help model alternative RNA structures and increased control over simulation parameters, enabling the simulations of conformational space searching, folding, and unfolding. [Dec. 2023]
- Enhanced data processing and analysis: Improved data processing capabilities, with a wider array of options for clustering folding trajectories and generating representative models, and reanalyzing the same simulation multiple times using different parameters, facilitating a more thorough exploration of RNA folding landscapes [Dec. 2023].
To download a standalone version of SimRNA, visit https://genesilico.pl/software/stand-alone/simrna/
References
SimRNAweb v2.0: A web server for RNA folding simulations and 3D structure modeling, with optional restraints and enhanced analysis of folding trajectories
S. Naeim Moafinejad, Belisa R H de Aquino, Michał J. Boniecki, Iswarya P N Pandaranadar Jeyeram, Grigory Nikolaev, Marcin Magnus2, Masoud Amiri Farsani, Nagendar Goud Badepally, Tomasz K. Wirecki, Filip Stefaniak, Janusz M. Bujnicki
Nucleic Acids Res 2024 [doi: 10.1093/nar/gkw279]
SimRNAweb: a web server for RNA 3D structure modeling with optional restraints
Magnus M*, Boniecki MJ*, Dawson W, Bujnicki JM
Nucleic Acids Res 2016 [doi: 10.1093/nar/gkae356]
SimRNA: a coarse-grained method for RNA folding simulations and 3D structure prediction
Boniecki MJ, Lach G, Dawson WK, Tomala K, Lukasz P, Soltysinski T, Rother KM, Bujnicki JM
Nucleic Acids Res 2015 [doi: 10.1093/nar/gkv1479]