RNArchitecture is a database that provides a comprehensive description of relationships between known families of structured non-coding RNAs, with a focus on structural similarities. The classification is hierarchical and similar to the system used in the SCOP and CATH databases of protein structures. Its central level is Family, which builds on the Rfam catalog, and gathers closely related RNAs. Consensus structures of Families are described with a reduced secondary structure representation. Evolutionarily related Families are grouped into Clans and Superfamilies. Similar structural motives are further grouped into Architectures. The highest level, Class, organizes Families into very broad structural categories, such as simple or complex structured RNAs, or lncRNA domains with unknown structure. Some groups at different levels of the hierarchy are currently labeled as "unclassified". The classification is expected to evolve as new data become available. For each Family with an experimentally determined 3D structure(s), a representative one is provided. RNArchitecture also presents theoretical models of RNA 3D structure and is open for submission of structural models by users. Compared to other databases, RNArchitecture is unique in its focusing on structure-based RNA classification, and in providing a platform for storing RNA 3D structural model information.
Maintaining the RNArchitecture database is a team effort of the current and past members of the Bujnicki Group. The members have helped with the manual curation of the database and some have developed algorithms to aid with the automated aspects of maintaining and updating it (like rna-pdb-tools, a toolbox to analyze structures and simulations of RNA).
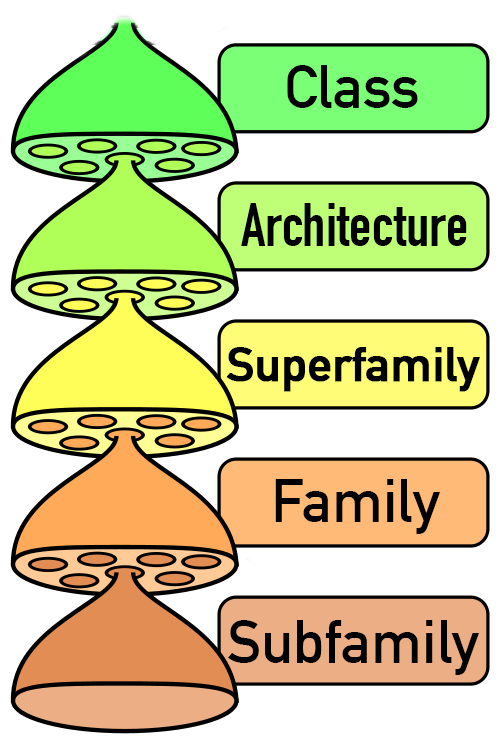
RNArchitecture starts with the Rfam-level classification of Families. This level of classification is mostly relevant from the point of view of molecular biology (RNA function). Families whose members exhibit structural variation are further subdivided into Subfamilies.
The database builds on Families to introduce the next level of Superfamily, grouping together all RNAs related by evolution. This category is used to group together close RNAs (such as members of the same Clan in Rfam) as well as very remote homologs (sequences that lost sequence similarity but retain at least partial structural similarity). One Superfamily can also group together sequences that exhibit similar sequence, structure, and function, due to a very strong convergence towards one particular minimum on the evolutionary fitness landscape -- however, we consider such cases -- especially in complex structures -- very rare, as they would normally appear as strong positives in homology searches. This level of classification is particularly relevant from the point of view of evolutionary biology (remotely related RNAs do not necessarily share the same function).
Beyond Superfamilies, the database introduces a level of Architecture which groups together RNAs with similar structure at the level of general shapes, ranging from simple ones (such as "one hairpin-like structure", "three-way junction", and "a pseudoknot with two helical regions"), to very complicated ones (such as "ribosomal RNA"). At this level, the classification is no longer restricted by biological relevance, but it focuses on structural aspects that may be more relevant from the point of view of physical chemistry and structure dynamics. Grouping together evolutionarily unrelated, but structurally similar RNAs, provides a convenient platform to study issues such as RNA folding (e.g., is one Architecture necessarily associated with the same folding pathway, regardless of its evolutionary origin). Furthermore, a structural analogy can be used in comparative modeling (e.g., if an evolutionarily related template is not available), to generate starting structure models for further optimization.
At the highest level, the database provides Classes, which group together structures according to the level of complexity rather than the actual shapes. This grouping can be practically relevant, for example, in biochemical or biophysical experiments that aim at targeting very broad classes or RNA molecules behaving similarly to each other under given experimental conditions, regardless of their biological function, (e.g., small structured RNAs vs long RNAs).
In the RNArchitecture database, the idea of "Clan" has been adapted from MEROPS and PFAM databases. Clans in Rfam describe related RNA families, which share sufficient sequence similarity to be detectable with sequence searches. The philosophy and methodology of RNArchitecture is different: in our database the level we initially called "Clans" is defined more similarly to the level of "Superfamily" in the SCOP database: in RNArchitecture this level is used to group families that are likely to be evolutionarily related, even if the relationship is very remote - e.g., in the complete absence of detectable sequence similarity and even if structural differences are observed.
Consequently, we decided to name "our Clans" a "Superfamilies", to distinguish them from "Rfam Clans", and to highlight the similarity to SCOP. In the current version of RNArchitecture database, Clans (from Rfam) are not included explicitly (except for being a part of Rfam annotation). Nonetheless, information about relationships between Families that are "clanned" together in Rfam is taken into account in our classification into Superfamilies: Rfam Families that belong to the same Rfam Clan essentially always belong to one Superfamily in our database. However, a given Superfamily often contains additional members (see the example below).
In the current release of RNArchitecture, we took into account the analysis by Lessa et al. (Genes 2012, 3(3), 378-390; doi:10.3390/genes3030378). They used a sequence clustering approach to detect remote homologies and suggested the grouping of many RNA families into clusters. This grouping has already been adopted to some extent in the last release of Rfam, and we have also taken into account the largest clusters reported in that work.
For example, our CRISPR-DR Superfamily includes the CRISPR-1 (CL00014) Rfam Clan, but includes not only CRISPR-DR2, CRISPR-DR4, CRISPR-DR14, CRISPR-DR17, CRISPR-DR25, CRISPR-DR43, and CRISPR-DR66 as in Rfam, but also other 57 families such as CRISPR-DR63, CRISPR-DR7, etc., as identified in sequence clustering by Lessa et al. (Genes 2012, 3(3), 378-390; doi:10.3390/genes3030378). Therefore, a Superfamily in RNArchitecture should be considered a higher level of classification (spanning bigger evolutionary distances) than Clans in Rfam. We envisage that the current catalog of Superfamilies will be extended and "isolated" Families will be included into existing Superfamilies, and new Superfamilies will be formed as new evolutionary relationships are discovered - exactly as it has been done over the years in the SCOP database.
Eggenhofer et al. (Nucleic Acids Res. 2013 Jul;41(Web Server issue):W499-503. doi: 10.1093/nar/gkt329) presented the CMCompare tool for family-family comparison. The CMCompare webserver compares Infernal RNA family models to identify models with poor specificity and explore the relationship between models. The CMCws provides options to compare new models against all existing models in the current Rfam database to avoid the construction of duplicate models for the same non-coding RNA family. In addition, the user can explore the relationship between two or more models, including whole sets of user-created family models. Authors have not proposed any particular clustering of Rfam families. We envisage that a result of all-vs-all clustering of Rfam families with CMCompare can be used to improve the classification of Families into Superfamilies in the next release of RNArchitecture.
Widmann et al. (RNA. 2012 Jul; 18(7): 1319–1327. doi: 10.1261/rna.032052.111) developed RNASTAR: An RNA Structural Alignment Repository. Authors manually curated a set of 148 alignments with a total of 9600 unique sequences, in which each alignment was backed by at least one crystal or NMR structure. These alignments included both naturally and artificially selected molecules. The improved alignments have been contributed to the Rfam database. We envisage that data from this resource (structure-assisted alignments) can be used in the future to refine the Family classification.
Izzo et al. (BMC Bioinformatics. 2011 May 31;12:219. doi: 10.1186/1471-2105-12-219) described RAG: RNA-As-Graphs database, which classified, catalogued, and predicted RNA secondary structure motifs using clustering and build-up approaches. It enables the search for novel RNA functionality - by classifying available motifs, suggesting new motifs for design, and allowing for more specific searches for specific topologies. The RAG web resource offers users a graph-based tool for exploring available RNA motifs and suggesting new RNAs for design.
. , & , ( ) , [ ] , { } , < >
If other characters are used, the search will return no results. For more information on sequence searching and available options, go to the help page.
([)]
()()
Yes! We are waiting for both - solved structures and generated models. Before deposition to the database, submitted structures will be evaluated. To deposit the structure, go to the contribution page and fill the form. We will contact you as soon as we process your application.
Please make sure that the model is properly refined with no clashscore and has a proper numbering and format. To check your model you can use MolProbity web server and to unify the format of your model you can use our software: rna-pdb-tools. For this, please use the command:
python rna_pdb_toolsx.py --get_rnapuzzle_ready input.pdb > output.pdb
Or, for batch processing:
for i in *.pdb; do rna_pdb_toolsx.py --get_rnapuzzle_ready $i > ${i/.pdb/_rpr.pdb}; done
The initial release contains 2688 RNA Families, 1738 Superfamilies, 18 Architectures, and 3 Classes. To see the summary of the current content of the database go to the status page. An interactive sunburst plot illustrates the hierarchy of RNArchitecture and the content of the current release.
RNArchitecture: a database and a classification system of RNA families, with a focus on structural information, Pietro Boccaletto, Marcin Magnus, Catarina Almeida, Adriana Żyła, Astha, Radosław Pluta, Błażej Bagiński, Elżbieta Jankowska, Stanisław Dunin-Horkawicz, Tomasz Wirecki, Michał Boniecki, Filip Stefaniak, Janusz M. Bujnicki [in review]
Please contact us!
Please see the help page or contact us.